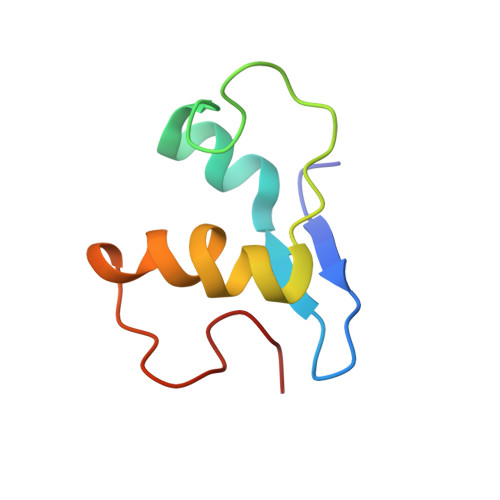
A proton-NMR investigation of the fully reduced cytochrome c7 from Desulfuromonas acetoxidans. Comparison between the reduced and the oxidized forms.
Assfalg, M., Banci, L., Bertini, I., Bruschi, M., Giudici-Orticoni, M.T.(1999) Eur J Biochem 266: 634-643
- PubMed: 10561607
- DOI: https://doi.org/10.1046/j.1432-1327.1999.00904.x
- Primary Citation of Related Structures:
1EHJ, 1F22 - PubMed Abstract:
The solution structure via 1H NMR of the fully reduced form of cytochrome c7 has been obtained. The protein sample was kept reduced by addition of catalytic amounts of Desulfovibrio gigas iron hydrogenase in H2 atmosphere after it had been checked that the presence of the hydrogenase did not affect the NMR spectrum. A final family of 35 conformers with rmsd values with respect to the mean structure of 8.7 +/- 1.5 nm and 12.4 +/- 1.3 nm for the backbone and heavy atoms, respectively, was obtained. A highly disordered loop involving residues 54-61 is present. If this loop is ignored, the rmsd values are 6.2 +/- 1.1 nm and 10.2 +/- 1.0 nm for the backbone and heavy atoms, respectively, which represent a reasonable resolution. The structure was analyzed and compared with the already available structure of the fully oxidized protein. Within the indetermination of the two solution structures, the result for the two redox forms is quite similar, confirming the special structural features of the three-heme cluster. A useful comparison can be made with the available crystal structures of cytochromes c3, which appear to be highly homologous except for the presence of a further heme. Finally, an analysis of the factors affecting the reduction potentials of the heme irons was performed, revealing the importance of net charges in differentiating the reduction potential when the other parameters are kept constant.
Organizational Affiliation:
Magnetic Resonance Center and Department of Chemistry, University of Florence, Italy.