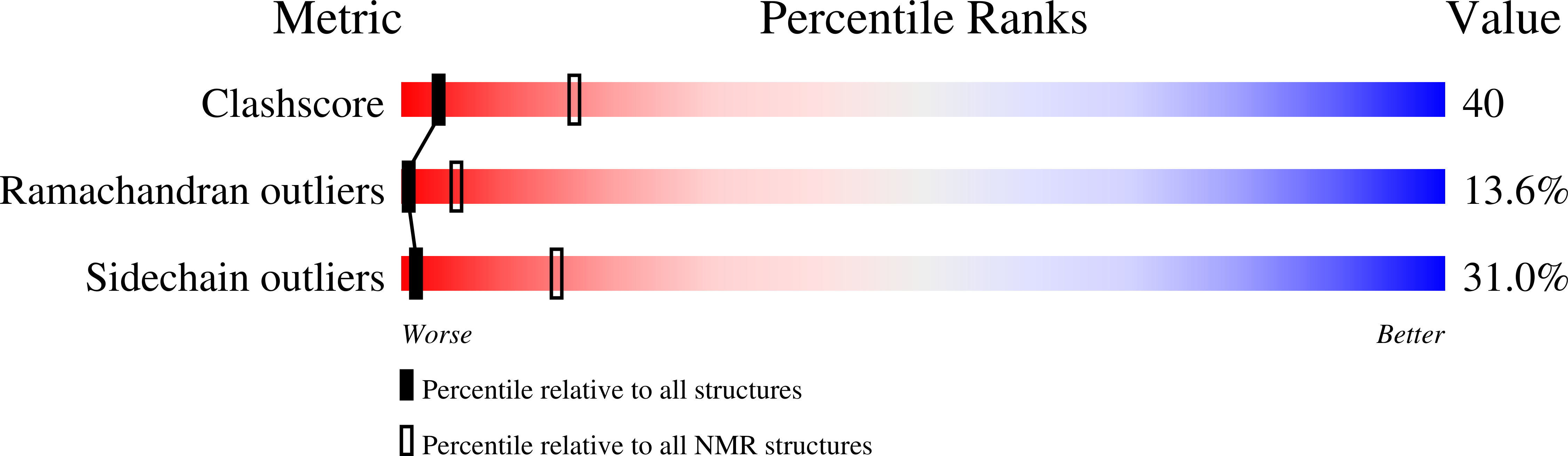Refined solution structure of the DNA-binding domain of GAL4 and use of 3J(113Cd,1H) in structure determination.
Baleja, J.D., Thanabal, V., Wagner, G.(1997) J Biomol NMR 10: 397-401
- PubMed: 9460244
- DOI: https://doi.org/10.1023/a:1018332327565
- Primary Citation of Related Structures:
1AW6 - PubMed Abstract:
We have refined the solution structure of cadmium-bound GAL4 and present its 15N and 1H NMR assignments. The root-mean-square (rms) deviation to the average structure was 0.4 +/- 0.05 A for backbone atoms, and 0.9 +/- 0.1 A for all heavy atoms. The three-bond heteronuclear 3J(113Cd,1H) coupling constants were found to disobey a Karplus-type relationship, which was attributable to the unusual constraints imposed by the bimetal-thiolate cluster in GAL4. We conclude that the structural parameters that correlate to 3J(113Cd,1H) are complex.
Organizational Affiliation:
Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115, USA.