Crystal structure of the kainate receptor GluR5 ligand-binding core in complex with (S)-glutamate
Naur, P., Vestergaard, B., Skov, L.K., Egebjerg, J., Gajhede, M., Kastrup, J.S.(2005) FEBS Lett 579: 1154-1160
- PubMed: 15710405
- DOI: https://doi.org/10.1016/j.febslet.2005.01.012
- Primary Citation of Related Structures:
1YCJ - PubMed Abstract:
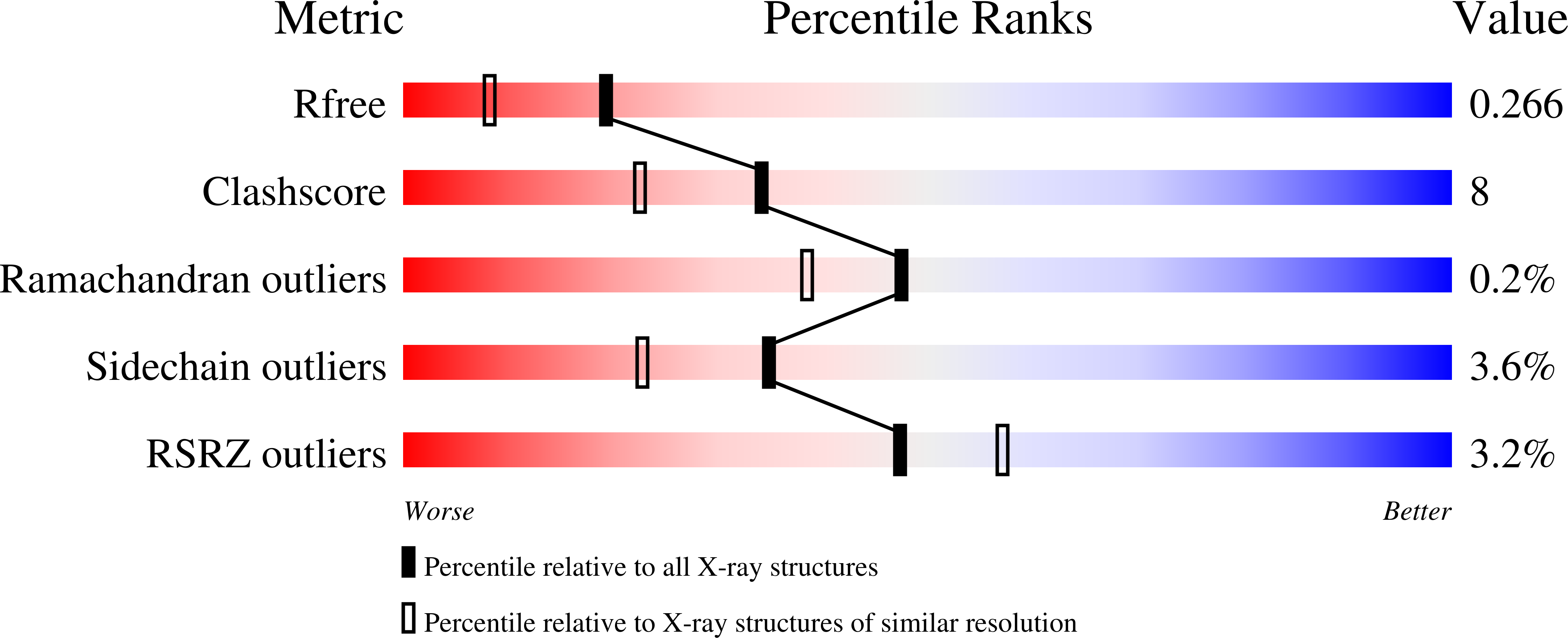
The X-ray structure of the ligand-binding core of the kainate receptor GluR5 (GluR5-S1S2) in complex with (S)-glutamate was determined to 1.95 A resolution. The overall GluR5-S1S2 structure comprises two domains and is similar to the related AMPA receptor GluR2-S1S2J. (S)-glutamate binds as in GluR2-S1S2J. Distinct features are observed for Ser741, which stabilizes a highly coordinated network of water molecules and forms an interdomain bridge. The GluR5 complex exhibits a high degree of domain closure (26 degrees) relative to apo GluR2-S1S2J. In addition, GluR5-S1S2 forms a novel dimer interface with a different arrangement of the two protomers compared to GluR2-S1S2J.
Organizational Affiliation:
Biostructural Research, Department of Medicinal Chemistry, Danish University of Pharmaceutical Sciences, Universitetsparken 2, DK-2100 Copenhagen, Denmark.