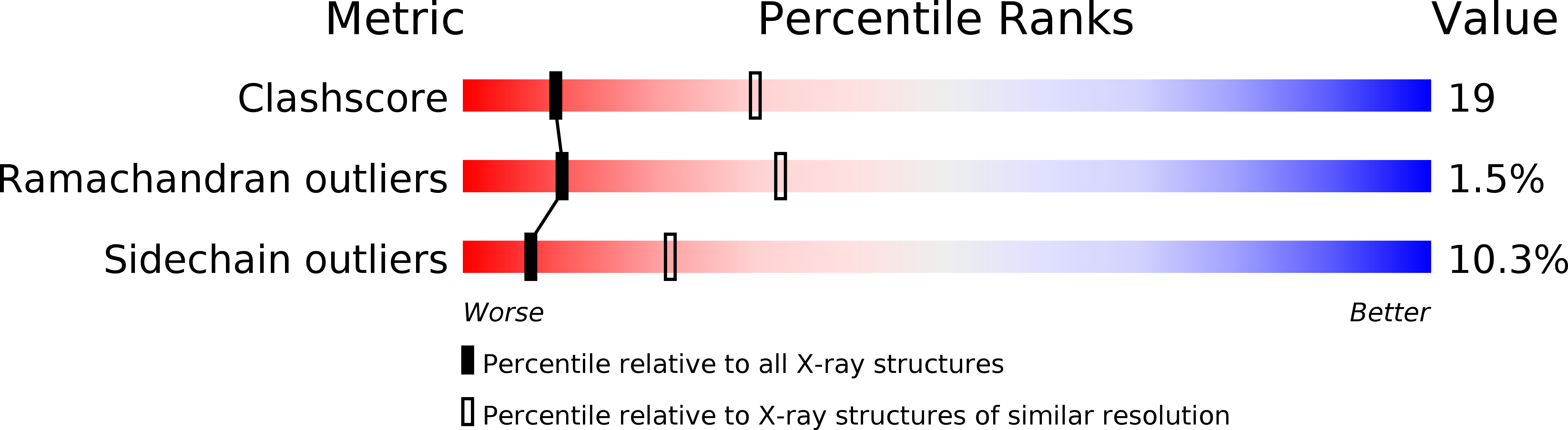Crystal structure of serine dehydratase from rat liver.
Yamada, T., Komoto, J., Takata, Y., Ogawa, H., Pitot, H.C., Takusagawa, F.(2003) Biochemistry 42: 12854-12865
- PubMed: 14596599
- DOI: https://doi.org/10.1021/bi035324p
- Primary Citation of Related Structures:
1PWE, 1PWH - PubMed Abstract:
SDH (L-serine dehydratase, EC 4.3.1.17) catalyzes the pyridoxal 5'-phosphate (PLP)-dependent dehydration of L-serine to yield pyruvate and ammonia. Liver SDH plays an important role in gluconeogenesis. Formation of pyruvate by SDH is a two-step reaction in which the hydroxyl group of serine is cleaved to produce aminoacrylate, and then the aminoacrylate is deaminated by nonenzymatic hydrolysis to produce pyruvate. The crystal structure of rat liver apo-SDH was determined by single isomorphous replacement at 2.8 A resolution. The holo-SDH crystallized with O-methylserine (OMS) was also determined at 2.6 A resolution by molecular replacement. SDH is composed of two domains, and each domain has a typical alphabeta-open structure. The active site is located in the cleft between the two domains. The holo-SDH contained PLP-OMS aldimine in the active site, indicating that OMS can form the Schiff base linkage with PLP, but the subsequent dehydration did not occur. Apo-SDH forms a dimer by inserting the small domain into the catalytic cleft of the partner subunit so that the active site is closed. Holo-SDH also forms a dimer by making contacts at the back of the clefts so that the dimerization does not close the catalytic cleft. The phosphate group of PLP is surrounded by a characteristic G-rich sequence ((168)GGGGL(172)) and forms hydrogen bonds with the amide groups of those amino acid residues, suggesting that the phosphate group can be protonated. N(1) of PLP participates in a hydrogen bond with Cys303, and similar hydrogen bonds with N(1) participating are seen in other beta-elimination enzymes. These hydrogen bonding schemes indicate that N(1) is not protonated, and thus, the pyridine ring cannot take a quinone-like structure. These characteristics of the bound PLP suggest that SDH catalysis is not facilitated by forming the resonance-stabilized structure of the PLP-Ser aldimine as seen in aminotransferases. A possible catalytic mechanism involves the phosphate group, surrounded by the characteristic sequence, acting as a general acid to donate a proton to the leaving hydroxyl group of serine.
Organizational Affiliation:
Department of Molecular Biosciences, University of Kansas, 1200 Sunnyside Avenue, Lawrence, Kansas 66045-7534, USA.