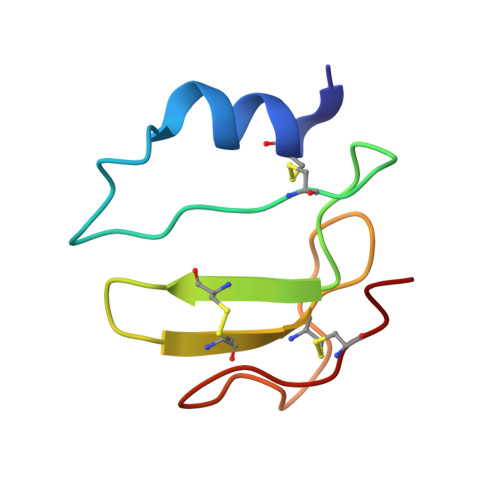
Synthesis and NMR structure of P41ICF, a potent inhibitor of human cathepsin L
Chiva, C., Barthe, P., Codina, A., Gairi, M., Molina, F., Granier, C., Pugniere, M., Inui, T., Nishio, H., Nishiuchi, Y., Kimura, T., Sakakibara, S., Albericio, F., Giralt, E.(2003) J Am Chem Soc 125: 1508-1517
- PubMed: 12568610
- DOI: https://doi.org/10.1021/ja0207908
- Primary Citation of Related Structures:
1L3H - PubMed Abstract:
The total synthesis and structural characterization of the MHCII-associated p41 invariant chain fragment (P41icf) is described. P41icf plays a crucial role in the maturation of MHC class II molecules and antigen processing, acting as a highly selective cathepsin L inhibitor. P41icf synthesis was achieved using a combined solid-phase/solution approach. The entire molecule (65 residues, 7246 Da unprotected) was assembled in solution from fully protected peptides in the size range of 10 residues. After deprotection, oxidative folding in carefully adjusted experimental conditions led to the completely folded and functional P41icf with a disulfide pairing identical to that of native P41icf. CD, NMR, and surface plasmon resonance (SPR) were used for the structural and functional characterization of synthetic P41icf. CD thermal denaturation showed clear cooperative behavior. Tight cathepsin L binding was demonstrated by SPR. (1)H NMR spectroscopy at 800 MHz of unlabeled P41icf was used to solve the three-dimensional structure of the molecule. P41icf behaves as a well-folded protein domain with a topology very close to the crystallographic cathepsin L-bound form.
Organizational Affiliation:
Institut de Recerca Biomèdica de Barcelona, Parc Científic de Barcelona, Spain.