Solution structure of Escherichia coli Par10: The prototypic member of the Parvulin family of peptidyl-prolyl cis/trans isomerases.
Kuehlewein, A., Voll, G., Alvarez, B.H., Kessler, H., Fischer, G., Rahfeld, J.U., Gemmecker, G.(2004) Protein Sci 13: 2378-2387
- PubMed: 15322281
- DOI: https://doi.org/10.1110/ps.04756704
- Primary Citation of Related Structures:
1JNS, 1JNT - PubMed Abstract:
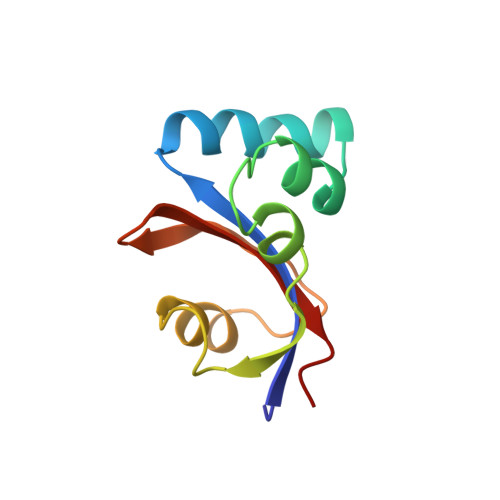
E. coli Par10 is a peptidyl-prolyl cis/trans isomerase (PPIase) from Escherichia coli catalyzing the isomerization of Xaa-Pro bonds in oligopeptides with a broad substrate specificity. The structure of E. coli Par10 has been determined by multidimensional solution-state NMR spectroscopy based on 1207 conformational constraints (1067 NOE-derived distances, 42 vicinal coupling-constant restraints, 30 hydrogen-bond restraints, and 68 phi/psi restraints derived from the Chemical Shift Index). Simulated-annealing calculations with the program ARIA and subsequent refinement with XPLOR yielded a set of 18 convergent structures with an average backbone RMSD from mean atomic coordinates of 0.50 A within the well-defined secondary structure elements. E. coli Par10 is the smallest known PPIase so far, with a high catalytic efficiency comparable to that of FKBPs and cyclophilins. The secondary structure of E. coli Par10 consists of four helical regions and a four-stranded antiparallel beta-sheet. The N terminus forms a beta-strand, followed by a large stretch comprising three alpha-helices. A loop region containing a short beta-strand separates these helices from a fourth alpha-helix. The C terminus consists of two more beta-strands completing the four-stranded anti-parallel beta-sheet with strand order 2143. Interestingly, the third beta-strand includes a Gly-Pro cis peptide bond. The curved beta-strand forms a hydrophobic binding pocket together with alpha-helix 4, which also contains a number of highly conserved residues. The three-dimensional structure of Par10 closely resembles that of the human proteins hPin1 and hPar14 and the plant protein Pin1At, belonging to the same family of highly homologous proteins.
Organizational Affiliation:
Department Chemie, OC II, TU München, Lichtenbergstr. 4, D-85747 Garching, Germany.