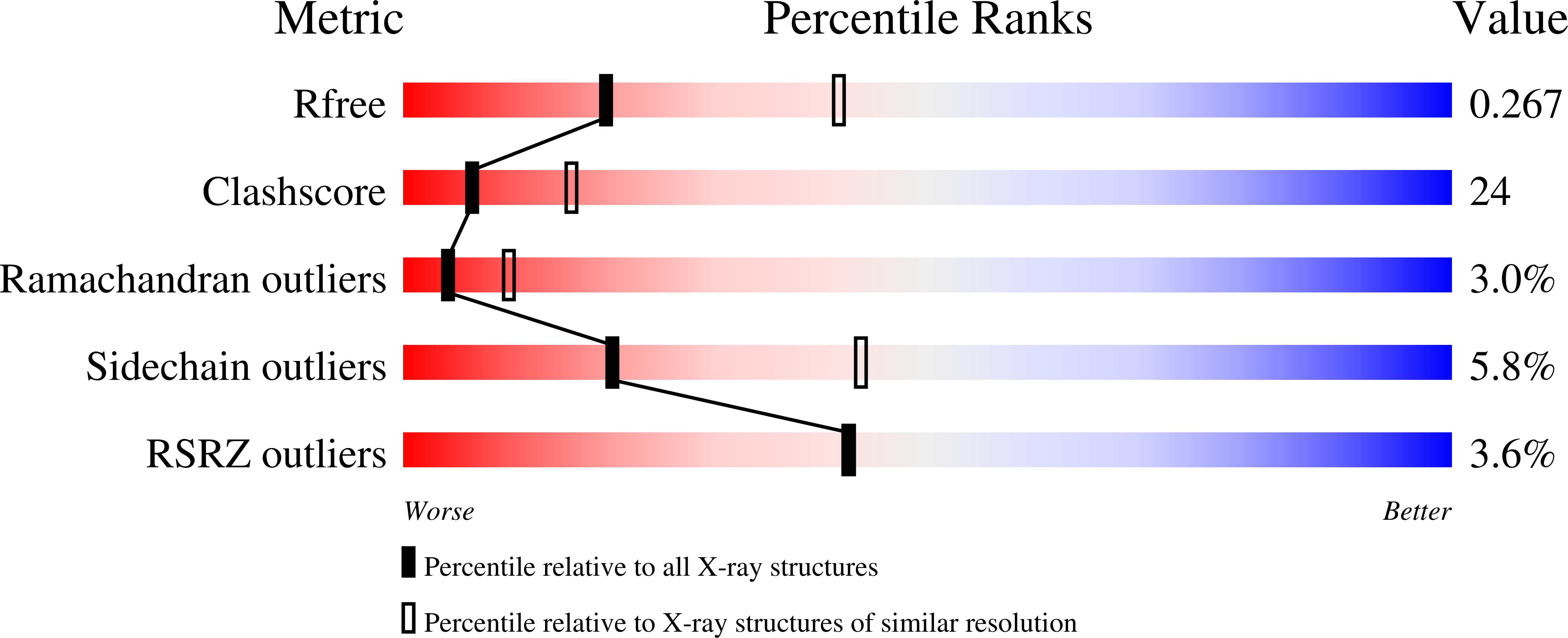
Structural basis of the redox switch in the OxyR transcription factor.
Choi, H., Kim, S., Mukhopadhyay, P., Cho, S., Woo, J., Storz, G., Ryu, S.(2001) Cell 105: 103-113
- PubMed: 11301006
- DOI: https://doi.org/10.1016/s0092-8674(01)00300-2
- Primary Citation of Related Structures:
1I69, 1I6A - PubMed Abstract:
The Escherichia coli OxyR transcription factor senses H2O2 and is activated through the formation of an intramolecular disulfide bond. Here we present the crystal structures of the regulatory domain of OxyR in its reduced and oxidized forms, determined at 2.7 A and 2.3 A resolutions, respectively. In the reduced form, the two redox-active cysteines are separated by approximately 17 A. Disulfide bond formation in the oxidized form results in a significant structural change in the regulatory domain. The structural remodeling, which leads to different oligomeric associations, accounts for the redox-dependent switch in OxyR and provides a novel example of protein regulation by "fold editing" through a reversible disulfide bond formation within a folded domain.
Organizational Affiliation:
Center for Cellular Switch Protein Structure, Korea Research Institute of Bioscience and, Biotechnology, P.O. Box 115, Yusong, 305-600, Taejon, South Korea