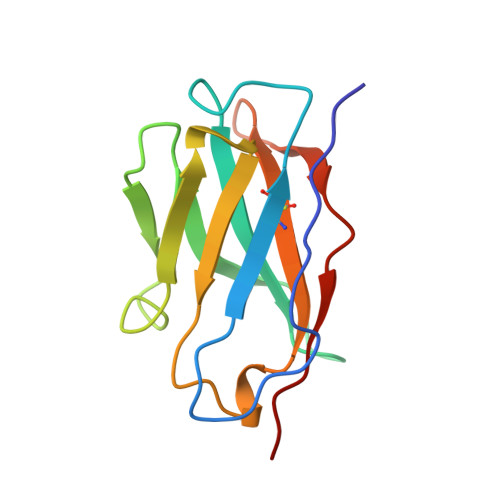
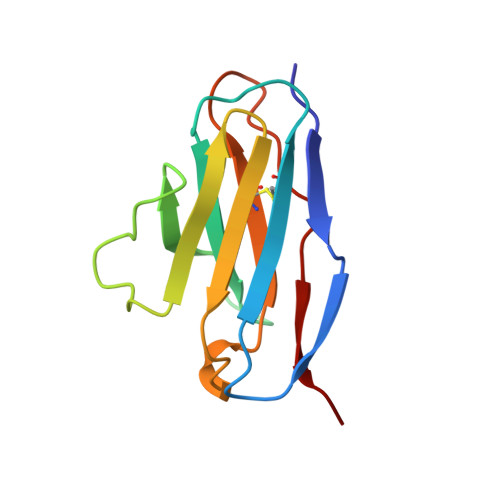
Structural consequences of humanizing an antibody.
Holmes, M.A., Foote, J.(1997) J Immunol 158: 2192-2201
- PubMed: 9036965
- Primary Citation of Related Structures:
1BVL - PubMed Abstract:
We have determined the crystal structure of the Fv fragment of a humanized anti-hen eggwhite lysozyme Ab (HuLys). This molecule is a composite of known structures: complementarity-determining regions (CDRs) were from the mouse anti-lysozyme Ab D1.3, heavy chain framework regions were from the human myeloma protein NEW, and the light chain framework regions were consensus sequences similar to the Bence Jones protein REI. HuLys crystallized in space group P4(3)2(1)2, with two Fv molecules in the crystallographic asymmetric unit. The structure was solved by molecular replacement, using a composite of the parent structures as a search model. The resolution of the structure was 2.87 A, with an R factor of 22%. There were several unanticipated structural similarities and differences between HuLys and the mouse and human structures. The framework regions of HuLys were as close or closer in conformation to D1.3 than to the NEW or REI protein, despite overwhelming sequence identity with the human framework regions. The effect was most pronounced at the CDR-framework junction, showing that the CDRs can induce structural changes in framework residues, having the net effect in HuLys of reconforming the framework into a conformation more like D1.3. In the combining site, the grafted CDRs retained conformational equilibria seen in D1.3, demonstrating that humanizing can be applied to Abs that bind Ags through isomeric equilibrium or induced fit mechanisms. In addition, HuLys showed systematic differences from D1.3 that resembled domain rotations reported for other Abs. However, the V(H)-V(L) interface itself was unaffected, and the apparent movement was actually caused by rearrangements distant from this surface.
Organizational Affiliation:
Division of Molecular Medicine, Fred Hutchinson Cancer Research Center, Seattle, WA 98104, USA.