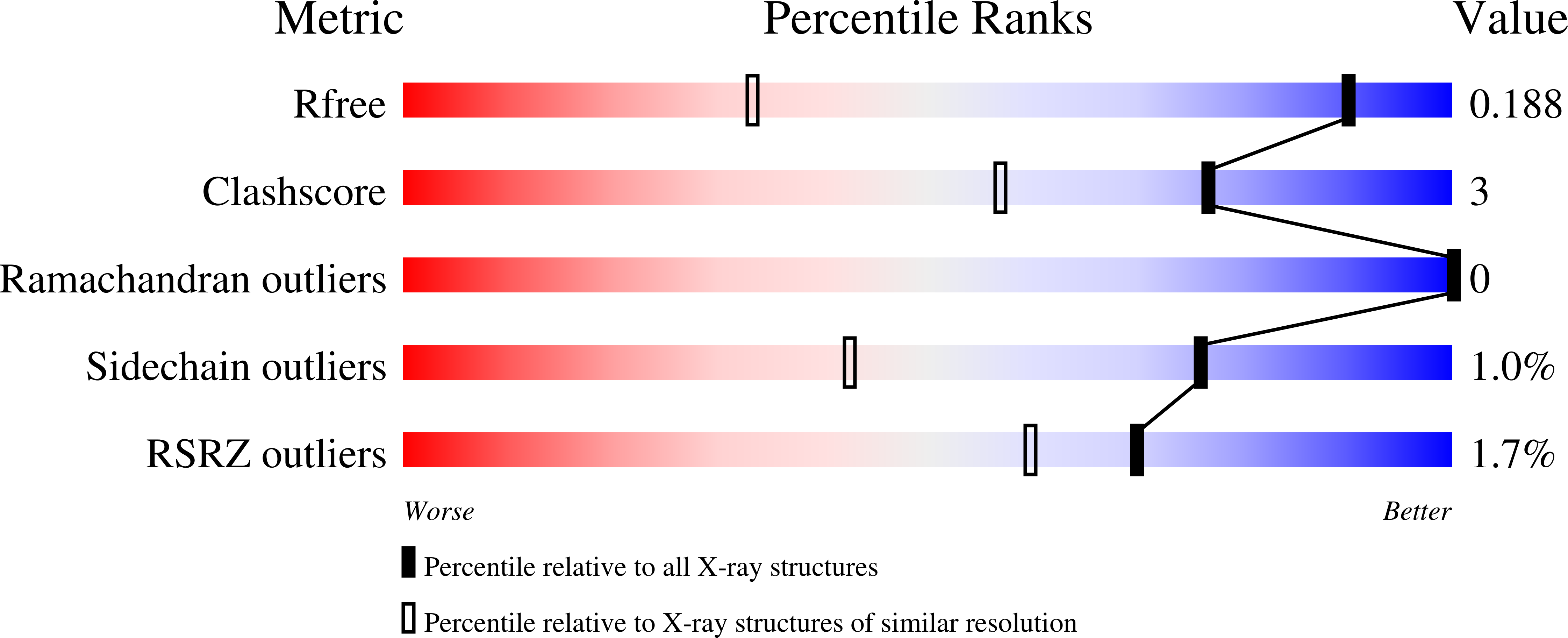
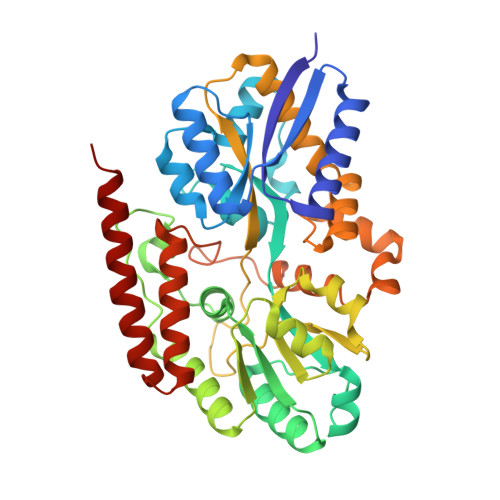
A metabolite binding protein moonlights as a bile-responsive chaperone.
Lee, C., Betschinger, P., Wu, K., Zyla, D.S., Glockshuber, R., Bardwell, J.C.(2020) EMBO J 39: e104231-e104231
- PubMed: 32882062
- DOI: https://doi.org/10.15252/embj.2019104231
- Primary Citation of Related Structures:
6X84 - PubMed Abstract:
Bile salts are secreted into the gastrointestinal tract to aid in the absorption of lipids. In addition, bile salts show potent antimicrobial activity in part by mediating bacterial protein unfolding and aggregation. Here, using a protein folding sensor, we made the surprising discovery that the Escherichia coli periplasmic glycerol-3-phosphate (G3P)-binding protein UgpB can serve, in the absence of its substrate, as a potent molecular chaperone that exhibits anti-aggregation activity against bile salt-induced protein aggregation. The substrate G3P, which is known to accumulate in the later compartments of the digestive system, triggers a functional switch between UgpB's activity as a molecular chaperone and its activity as a G3P transporter. A UgpB mutant unable to bind G3P is constitutively active as a chaperone, and its crystal structure shows that it contains a deep surface groove absent in the G3P-bound wild-type UgpB. Our work illustrates how evolution may be able to convert threats into signals that first activate and then inactivate a chaperone at the protein level in a manner that bypasses the need for ATP.
Organizational Affiliation:
Department of Molecular, Cellular, and Developmental Biology, Howard Hughes Medical Institute, University of Michigan, Ann Arbor, MI, USA.