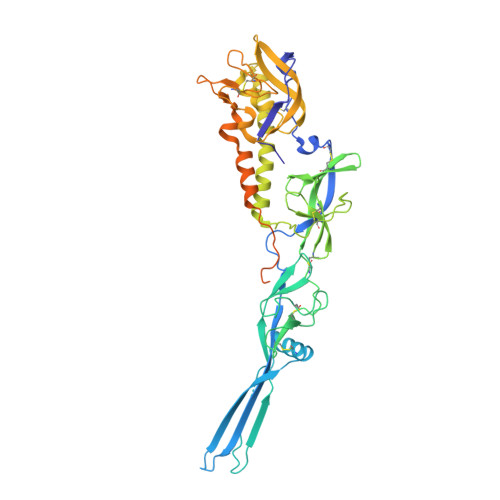
Crystal structure of Mokola virus glycoprotein in its post-fusion conformation.
Belot, L., Ouldali, M., Roche, S., Legrand, P., Gaudin, Y., Albertini, A.A.(2020) PLoS Pathog 16: e1008383-e1008383
- PubMed: 32150590
- DOI: https://doi.org/10.1371/journal.ppat.1008383
- Primary Citation of Related Structures:
6TMR - PubMed Abstract:
Mokola virus (MOKV) belongs to the lyssavirus genus. As other genus members-including rabies virus (RABV)-it causes deadly encephalitis in mammals. MOKV entry into host cells is mediated by its transmembrane glycoprotein G. First, G binds cellular receptors, triggering virion endocytosis. Then, in the acidic endosomal environment, G undergoes a conformational change from its pre- toward its post-fusion state that catalyzes the merger of the viral and endosomal membranes. Here, we have determined the crystal structure of a soluble MOKV G ectodomain in which the hydrophobic fusion loops have been replaced by more hydrophilic sequences. The crystal structure corresponds to a monomer that is similar to the protomer of the trimeric post-fusion state of vesicular stomatitis virus (VSV) G. However, by electron microscopy, we show that, at low pH, at the surface of pseudotyped VSV, MOKV spikes adopt the trimeric post-fusion conformation and have a tendency to reorganize into regular arrays. Sequence alignment between MOKV G and RABV G allows a precise location of RABV G antigenic sites. Repositioning MOKV G domains on VSV G pre-fusion structure reveals that antigenic sites are located in the most exposed part of the molecule in its pre-fusion conformation and are therefore very accessible to antibodies. Furthermore, the structure allows the identification of pH-sensitive molecular switches. Specifically, the long helix, which constitutes the core of the post-fusion trimer for class III fusion glycoproteins, contains many acidic residues located at the trimeric interface. Several of them, aligned along the helix, point toward the trimer axis. They have to be protonated for the post-fusion trimer to be stable. At high pH, when they are negatively charged, they destabilize the interface, which explains the conformational change reversibility. Finally, the present structure will be of great help to perform rational mutagenesis on lyssavirus glycoproteins.
Organizational Affiliation:
Institute for Integrative Biology of the Cell (I2BC), CEA, CNRS, Univ. Paris-Sud, Université Paris-Saclay, France.