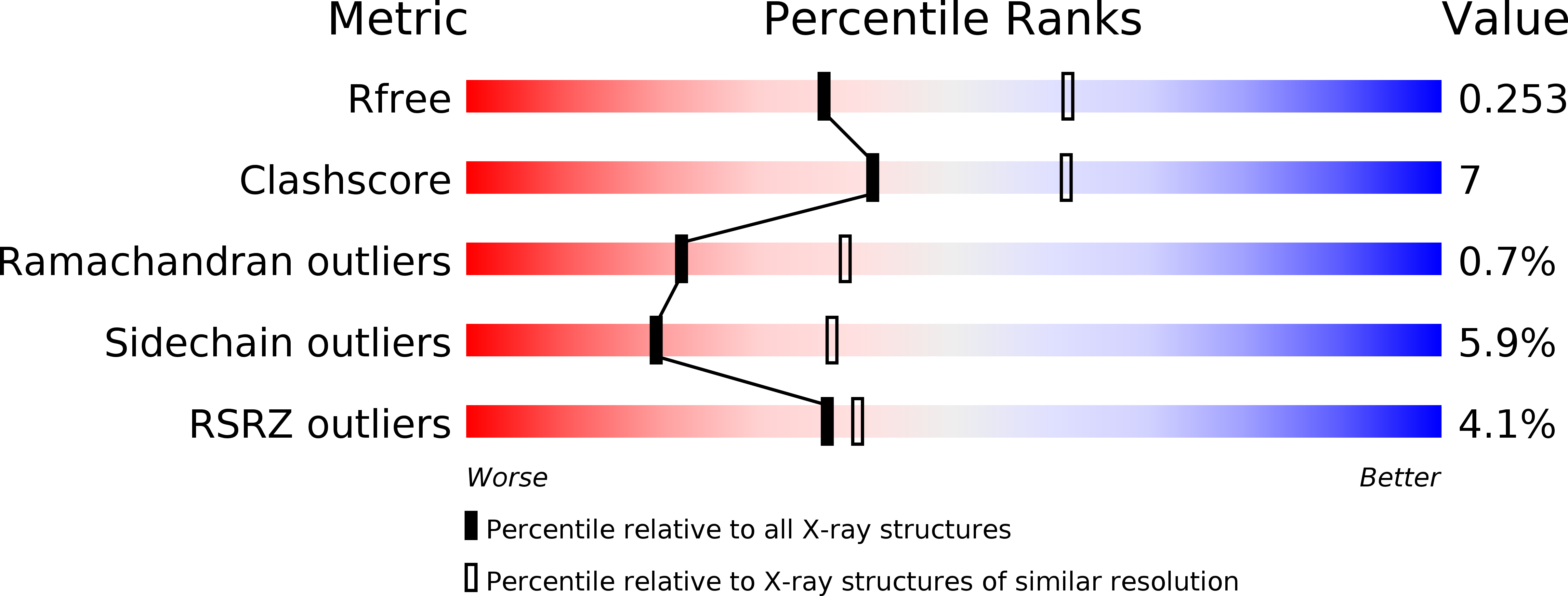
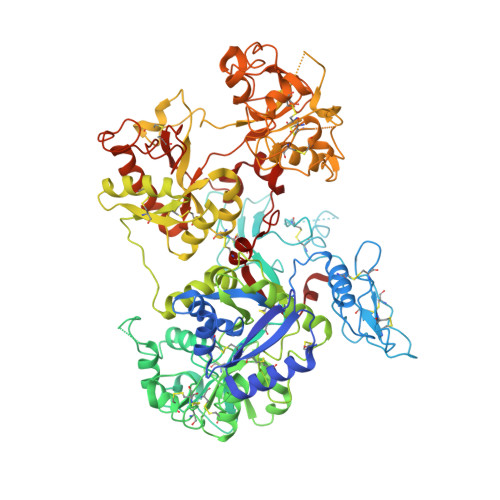
Structure of the saxiphilin:saxitoxin (STX) complex reveals a convergent molecular recognition strategy for paralytic toxins.
Yen, T.J., Lolicato, M., Thomas-Tran, R., Du Bois, J., Minor Jr., D.L.(2019) Sci Adv 5: eaax2650-eaax2650
- PubMed: 31223657
- DOI: https://doi.org/10.1126/sciadv.aax2650
- Primary Citation of Related Structures:
6O0D, 6O0E, 6O0F - PubMed Abstract:
Dinoflagelates and cyanobacteria produce saxitoxin (STX), a lethal bis-guanidinium neurotoxin causing paralytic shellfish poisoning. A number of metazoans have soluble STX-binding proteins that may prevent STX intoxication. However, their STX molecular recognition mechanisms remain unknown. Here, we present structures of saxiphilin (Sxph), a bullfrog high-affinity STX-binding protein, alone and bound to STX. The structures reveal a novel high-affinity STX-binding site built from a "proto-pocket" on a transferrin scaffold that also bears thyroglobulin domain protease inhibitor repeats. Comparison of Sxph and voltage-gated sodium channel STX-binding sites reveals a convergent toxin recognition strategy comprising a largely rigid binding site where acidic side chains and a cation-π interaction engage STX. These studies reveal molecular rules for STX recognition, outline how a toxin-binding site can be built on a naïve scaffold, and open a path to developing protein sensors for environmental STX monitoring and new biologics for STX intoxication mitigation.
Organizational Affiliation:
Cardiovascular Research Institute, University of California, San Francisco, San Francisco, CA 94158, USA.