Revisiting biomolecular NMR spectroscopy for promoting small-molecule drug discovery.
Hanzawa, H., Shimada, T., Takahashi, M., Takahashi, H.(2020) J Biomol NMR 74: 501-508
- PubMed: 32306215
- DOI: https://doi.org/10.1007/s10858-020-00314-0
- Primary Citation of Related Structures:
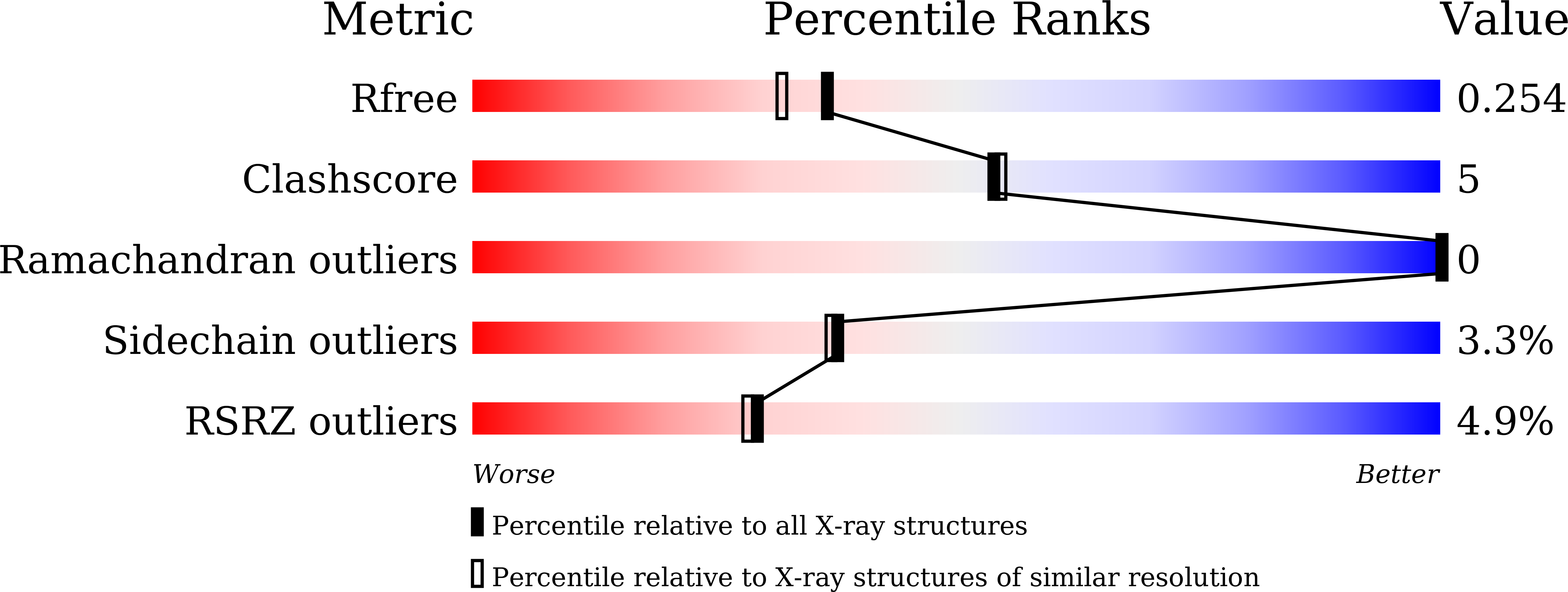
6LUR - PubMed Abstract:
Recently, there has been increasing interest in new modalities such as therapeutic antibodies and gene therapy at a number of pharmaceutical companies. Moreover, in small-molecule drug discovery at such companies, efforts have focused on hard-to-drug targets such as inhibiting protein-protein interactions. Biomolecular NMR spectroscopy has been used in drug discovery in a variety of ways, such as for the reliable detection of binding and providing three-dimensional structural information for structure-based drug design. The advantages of using NMR spectroscopy have been known for decades (Jahnke in J Biomol NMR 39:87-90, (2007); Gossert and Jahnke in Prog Nucl Magn Reson Spectrosc 97:82-125, (2016)). For tackling hard-to-drug targets and increasing the success in discovering drug molecules, in-depth analysis of drug-target protein interactions performed by biophysical methods will be more and more essential. Here, we review the advantages of NMR spectroscopy as a key technology of biophysical methods and also discuss issues such as using cutting-edge NMR spectrometers and increasing the demand of utilizing conformational dynamics information for promoting small-molecule drug discovery.
Organizational Affiliation:
Structure-Based Drug Design Group, Organic Synthesis Department, Daiichi Sankyo RD Novare Co., Ltd, 1-16-13 Kita-Kasai, Edogawa-ku, Tokyo, 134-8630, Japan. hanzawa.hiroyuki.ac@rdn.daiichisankyo.co.jp.