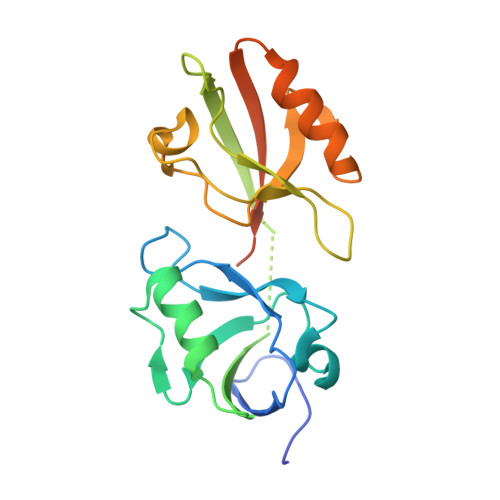
An unexpected INAD PDZ tandem-mediated plc beta binding in Drosophila photo receptors.
Ye, F., Huang, Y., Li, J., Ma, Y., Xie, C., Liu, Z., Deng, X., Wan, J., Xue, T., Liu, W., Zhang, M.(2018) Elife 7
- PubMed: 30526850
- DOI: https://doi.org/10.7554/eLife.41848
- Primary Citation of Related Structures:
6IRB, 6IRC, 6IRD, 6IRE - PubMed Abstract:
INAD assembles key enzymes of the Drosophila compound eye photo-transduction pathway into a supramolecular complex, supporting efficient and fast light signaling. However, the molecular mechanism that governs the interaction between INAD and NORPA (phospholipase Cβ, PLCβ), a key step for the fast kinetics of the light signaling, is not known. Here, we show that the NORPA C-terminal coiled-coil domain and PDZ-binding motif (CC-PBM) synergistically bind to INAD PDZ45 tandem with an unexpected mode and unprecedented high affinity. Guided by the structure of the INAD-NORPA complex, we discover that INADL is probably a mammalian counterpart of INAD. The INADL PDZ89 tandem specifically binds to PLCβ4 with a mode that is strikingly similar to that of the INAD-NORPA complex, as revealed by the structure of the INADL PDZ89-PLCβ4 CC-PBM complex. Therefore, our study suggests that the highly specific PDZ tandem - PLCβ interactions are an evolutionarily conserved mechanism in PLCβ signaling in the animal kingdom.
Organizational Affiliation:
Division of Life Science, State Key Laboratory of Molecular Neuroscience, Hong Kong University of Science and Technology, Hong Kong, China.