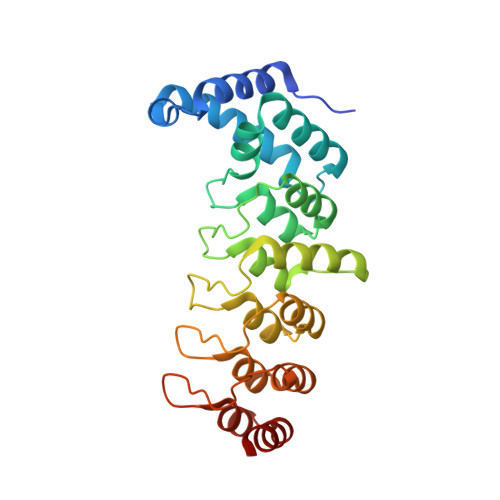
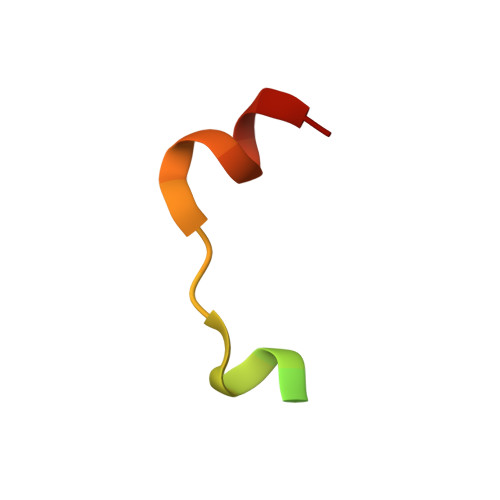
Structural insights into ankyrin repeat-mediated recognition of the kinesin motor protein KIF21A by KANK1, a scaffold protein in focal adhesion.
Pan, W., Sun, K., Tang, K., Xiao, Q., Ma, C., Yu, C., Wei, Z.(2018) J Biol Chem 293: 1944-1956
- PubMed: 29217769
- DOI: https://doi.org/10.1074/jbc.M117.815779
- Primary Citation of Related Structures:
5YAY, 5YAZ - PubMed Abstract:
Kidney ankyrin repeat-containing proteins (KANK1/2/3/4) belong to a family of scaffold proteins, playing critical roles in cytoskeleton organization, cell polarity, and migration. Mutations in KANK proteins are implicated in cancers and genetic diseases, such as nephrotic syndrome. KANK proteins can bind various target proteins through different protein regions, including a highly conserved ankyrin repeat domain (ANKRD). However, the molecular basis for target recognition by the ANKRD remains elusive. In this study, we solved a high-resolution crystal structure of the ANKRD of KANK1 in complex with a short sequence of the motor protein kinesin family member 21A (KIF21A), revealing that the highly specific target-binding mode of the ANKRD involves combinatorial use of two interfaces. Mutations in either interface disrupted the KANK1-KIF21A interaction. Cellular immunofluorescence localization analysis indicated that binding-deficient mutations block recruitment of KIF21A to focal adhesions by KANK1. In conclusion, our structural study provides mechanistic explanations for the ANKRD-mediated recognition of KIF21A and for many disease-related mutations identified in human KANK proteins.
Organizational Affiliation:
From the Department of Biology, Southern University of Science and Technology, 518055 Shenzhen, China.