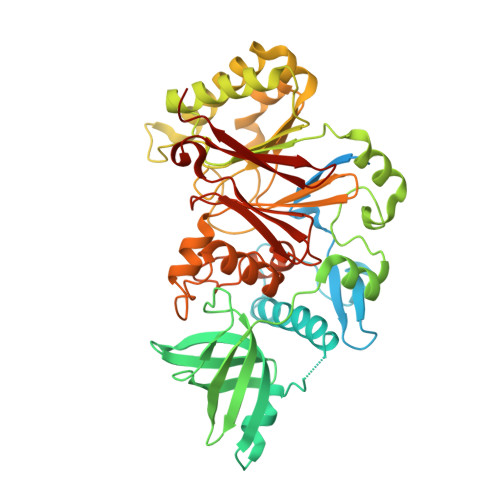
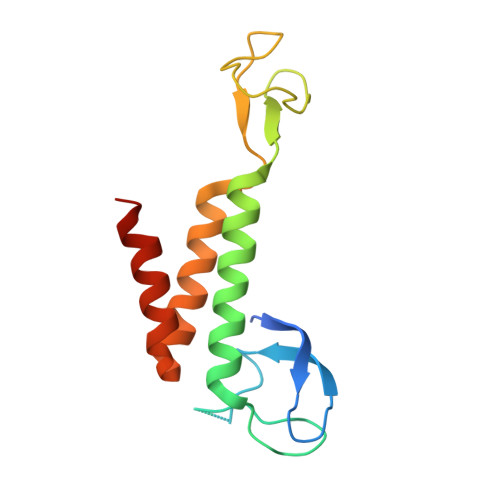
Crystal structure of the human Pol B-subunit in complex with the C-terminal domain of the catalytic subunit.
Baranovskiy, A.G., Gu, J., Babayeva, N.D., Kurinov, I., Pavlov, Y.I., Tahirov, T.H.(2017) J Biol Chem 292: 15717-15730
- PubMed: 28747437
- DOI: https://doi.org/10.1074/jbc.M117.792705
- Primary Citation of Related Structures:
5VBN - PubMed Abstract:
The eukaryotic B-family DNA polymerases include four members: Polα, Polδ, Polϵ, and Polζ, which share common architectural features, such as the exonuclease/polymerase and C-terminal domains (CTDs) of catalytic subunits bound to indispensable B-subunits, which serve as scaffolds that mediate interactions with other components of the replication machinery. Crystal structures for the B-subunits of Polα and Polδ/Polζ have been reported: the former within the primosome and separately with CTD and the latter with the N-terminal domain of the C-subunit. Here we present the crystal structure of the human Polϵ B-subunit (p59) in complex with CTD of the catalytic subunit (p261 C ). The structure revealed a well defined electron density for p261 C and the phosphodiesterase and oligonucleotide/oligosaccharide-binding domains of p59. However, electron density was missing for the p59 N-terminal domain and for the linker connecting it to the phosphodiesterase domain. Similar to Polα, p261 C of Polϵ contains a three-helix bundle in the middle and zinc-binding modules on each side. Intersubunit interactions involving 11 hydrogen bonds and numerous hydrophobic contacts account for stable complex formation with a buried surface area of 3094 Å 2 Comparative structural analysis of p59-p261 C with the corresponding Polα complex revealed significant differences between the B-subunits and CTDs, as well as their interaction interfaces. The B-subunit of Polδ/Polζ also substantially differs from B-subunits of either Polα or Polϵ. This work provides a structural basis to explain biochemical and genetic data on the importance of B-subunit integrity in replisome function in vivo .
Organizational Affiliation:
From the Eppley Institute for Research in Cancer and Allied Diseases, Fred & Pamela Buffett Cancer Center and.