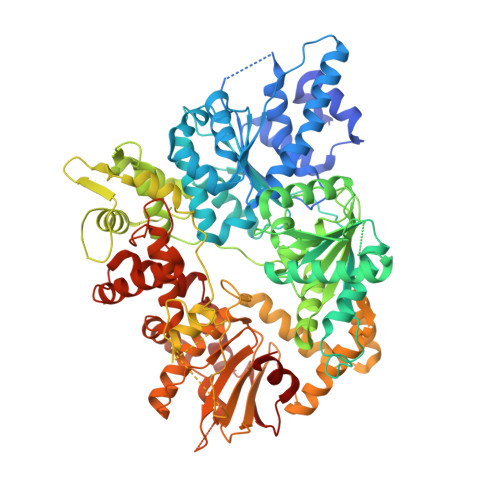
Structure of mycobacterial 3'-to-5' RNA:DNA helicase Lhr bound to a ssDNA tracking strand highlights distinctive features of a novel family of bacterial helicases.
Ejaz, A., Ordonez, H., Jacewicz, A., Ferrao, R., Shuman, S.(2018) Nucleic Acids Res 46: 442-455
- PubMed: 29165676
- DOI: https://doi.org/10.1093/nar/gkx1163
- Primary Citation of Related Structures:
5V9X - PubMed Abstract:
Mycobacterial Lhr is a DNA damage-inducible superfamily 2 helicase that uses adenosine triphosphate (ATP) hydrolysis to drive unidirectional 3'-to-5' translocation along single-stranded DNA (ssDNA) and to unwind RNA:DNA duplexes en route. ATPase, translocase and helicase activities are encompassed within the N-terminal 856-amino acid segment. The crystal structure of Lhr-(1-856) in complex with AMPPNP•Mg2+ and ssDNA defines a new helicase family. The enzyme comprises two N-terminal RecA-like modules, a winged helix (WH) domain and a unique C-terminal domain. The 3' ssDNA end binds in a crescent-shaped groove at the interface between the first RecA domain and the WH domain and tracks 5' into a groove between the second RecA and C domains. A kissing interaction between the second RecA and C domains forms an aperture that demarcates a putative junction between the loading strand tail and the duplex, with the first duplex nucleoside bookended by stacking on Trp597. Intercalation of Ile528 between nucleosides of the loading strand creates another bookend. Coupling of ATP hydrolysis to RNA:DNA unwinding is dependent on Trp597 and Ile528, and on Thr145 and Arg279 that contact phosphates of the loading strand. The structural and functional data suggest a ratchet mechanism of translocation and unwinding coupled to ATP-driven domain movements.
Organizational Affiliation:
Molecular Biology Program, Memorial Sloan Kettering Cancer Center, New York, NY 10065, USA.