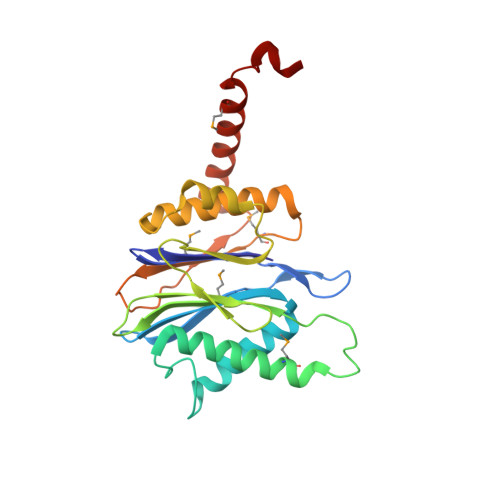
The Architecture of the Anbu Complex Reflects an Evolutionary Intermediate at the Origin of the Proteasome System.
Fuchs, A.C.D., Alva, V., Maldoner, L., Albrecht, R., Hartmann, M.D., Martin, J.(2017) Structure 25: 834-845.e5
- PubMed: 28479063
- DOI: https://doi.org/10.1016/j.str.2017.04.005
- Primary Citation of Related Structures:
5LOX, 5LOY - PubMed Abstract:
Proteasomes are self-compartmentalizing proteases that function at the core of the cellular protein degradation machinery in eukaryotes, archaea, and some bacteria. Although their evolutionary history is under debate, it is thought to be linked to that of the bacterial protease HslV and the hypothetical bacterial protease Anbu (ancestral beta subunit). Here, together with an extensive bioinformatic analysis, we present the first biophysical characterization of Anbu. Anbu forms a dodecameric complex with a unique architecture that was only accessible through the combination of X-ray crystallography and small-angle X-ray scattering. While forming continuous helices in crystals and electron microscopy preparations, refinement of sections from the crystal structure against the scattering data revealed a helical open-ring structure in solution, contrasting the ring-shaped structures of proteasome and HslV. Based on this primordial architecture and exhaustive sequence comparisons, we propose that Anbu represents an ancestral precursor at the origin of self-compartmentalization.
Organizational Affiliation:
Department of Protein Evolution, Max Planck Institute for Developmental Biology, Spemannstraße 35, 72076 Tübingen, Germany.