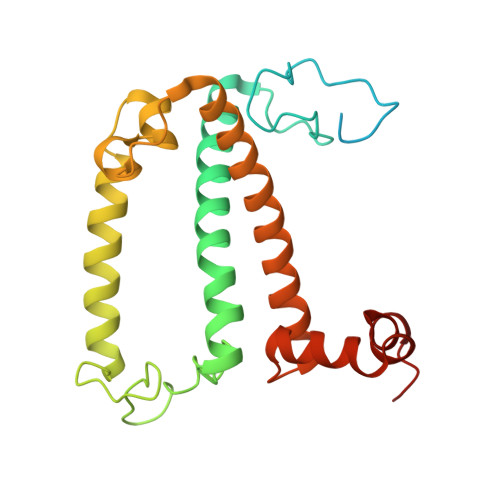
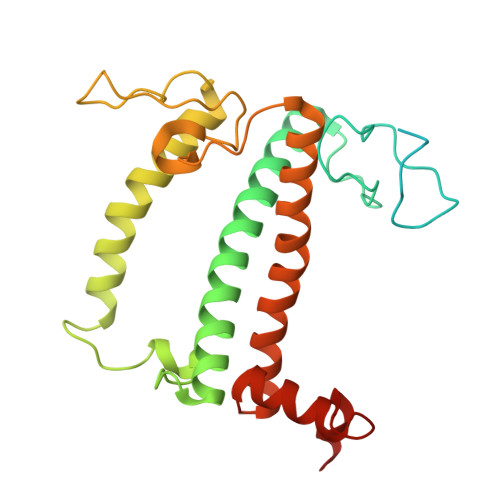
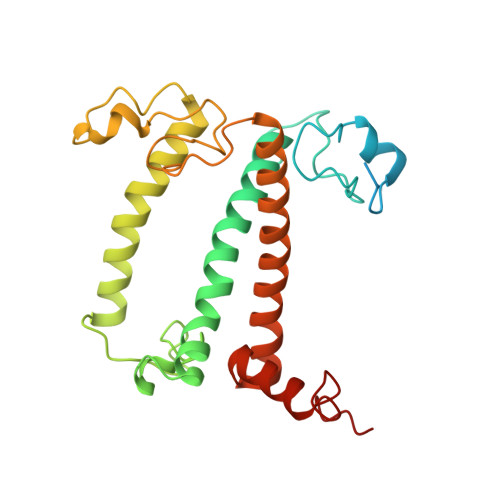
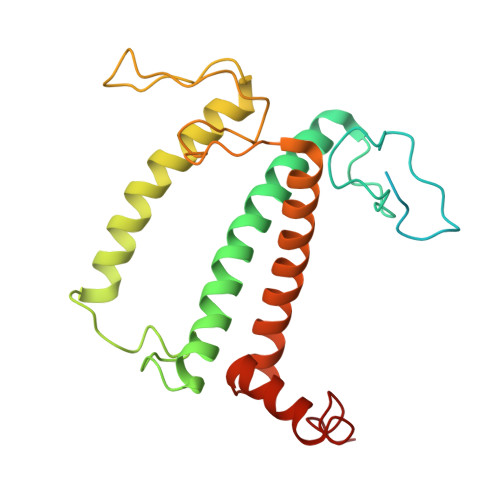
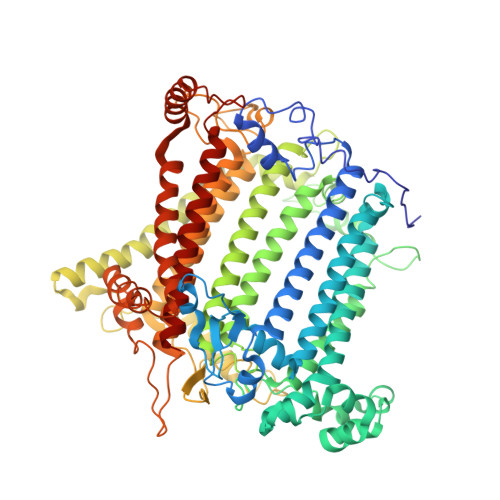
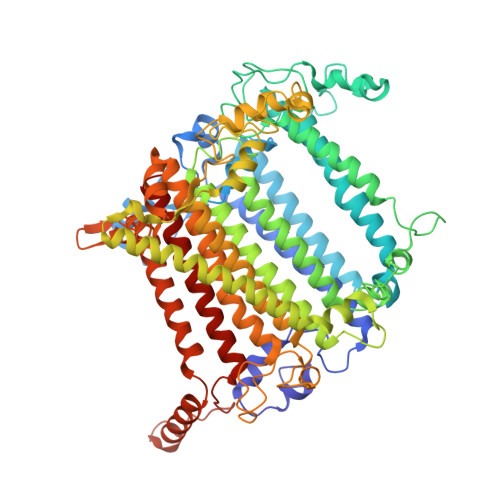
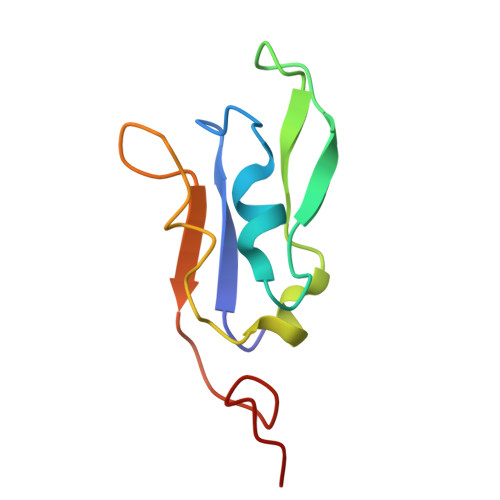
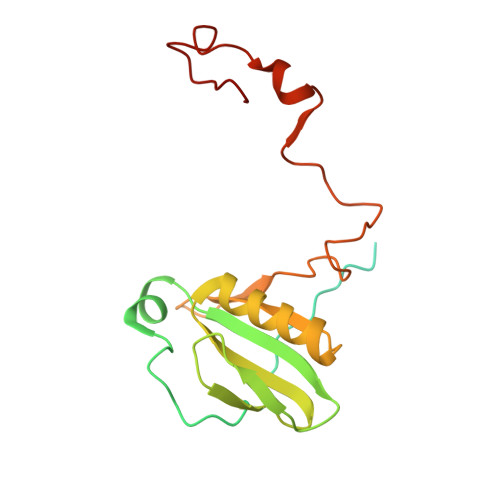
Structure of the maize photosystem I supercomplex with light-harvesting complexes I and II.
Pan, X., Ma, J., Su, X., Cao, P., Chang, W., Liu, Z., Zhang, X., Li, M.(2018) Science 360: 1109-1113
- PubMed: 29880686
- DOI: https://doi.org/10.1126/science.aat1156
- Primary Citation of Related Structures:
5ZJI - PubMed Abstract:
Plants regulate photosynthetic light harvesting to maintain balanced energy flux into photosystems I and II (PSI and PSII). Under light conditions favoring PSII excitation, the PSII antenna, light-harvesting complex II (LHCII), is phosphorylated and forms a supercomplex with PSI core and the PSI antenna, light-harvesting complex I (LHCI). Both LHCI and LHCII then transfer excitation energy to the PSI core. We report the structure of maize PSI-LHCI-LHCII solved by cryo-electron microscopy, revealing the recognition site between LHCII and PSI. The PSI subunits PsaN and PsaO are observed at the PSI-LHCI interface and the PSI-LHCII interface, respectively. Each subunit relays excitation to PSI core through a pair of chlorophyll molecules, thus revealing previously unseen paths for energy transfer between the antennas and the PSI core.
Organizational Affiliation:
National Laboratory of Biomacromolecules, CAS Center for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, P.R. China.