Proline iminopeptidase from Psychrophilic yeast glaciozyma antarctica
Kamaruddin, S., Rodzli, N.A., Jonet, A., Seman, W.M.K.W., Tab, M.M., Minor, N., Jaafar, N.R., Mahadi, N.M., Murad, A.M.A., Bakar, F.D.A.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cold active proline iminopeptidase | 355 | Glaciozyma antarctica | Mutation(s): 0 EC: 3.4.11.5 | 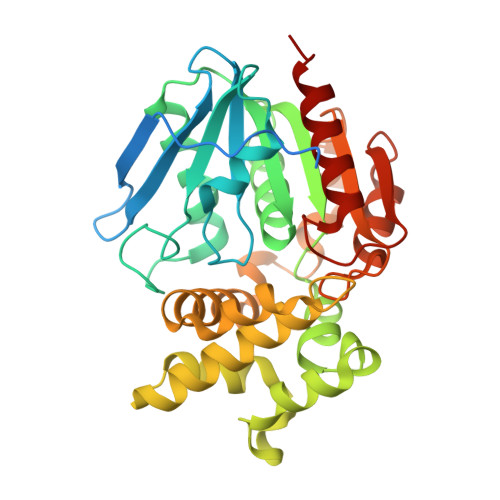 | |
UniProt | |||||
Find proteins for A0A2H4A311 (Glaciozyma antarctica) Explore A0A2H4A311 Go to UniProtKB: A0A2H4A311 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A2H4A311 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| FLC Query on FLC | C [auth A], D [auth A], E [auth B] | CITRATE ANION C6 H5 O7 KRKNYBCHXYNGOX-UHFFFAOYSA-K |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 55.388 | α = 90 |
| b = 82.269 | β = 90 |
| c = 171.005 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-3000 | data reduction |
| HKL-3000 | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| MINISTRY OF SCIENCE, TECHNOLOGY AND INNOVATION MALAYSIA | Malaysia | 02-05-20-SF0007 |