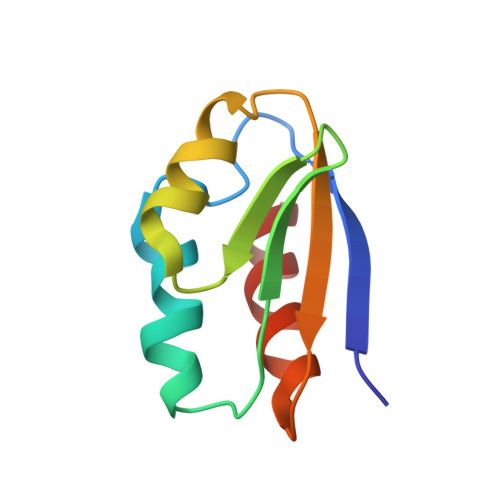
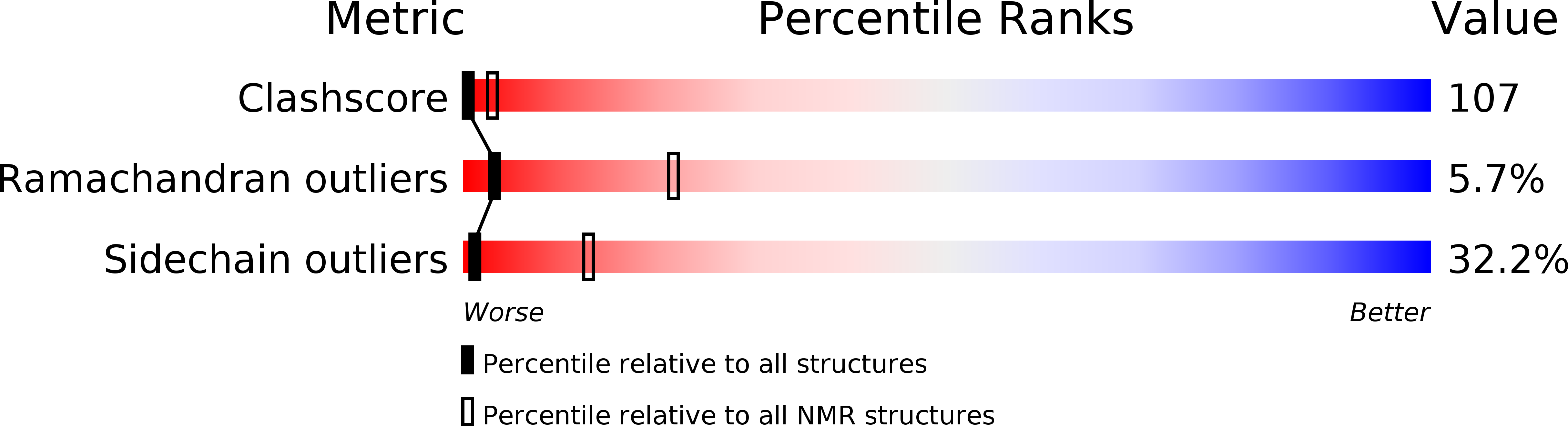Solution structure of NPr, a bacterial signal-transducing protein that controls the phosphorylation state of the potassium transporter-regulating protein IIA Ntr.
Li, X., Peterkofsky, A., Wang, G.(2008) Amino Acids 35: 531-539
- PubMed: 18421563
- DOI: https://doi.org/10.1007/s00726-008-0079-9
- Primary Citation of Related Structures:
5T17 - PubMed Abstract:
A nitrogen-related signal transduction pathway, consisting of the three phosphotransfer proteins EI(Ntr), NPr, and IIA(Ntr), was discovered recently to regulate the uptake of K(+) in Escherichia coli. In particular, dephosphorylated IIA(Ntr) inhibits the activity of the K(+) transporter TrkA. Since the phosphorylation state of IIA(Ntr) is partially determined by its reversible phosphorylation by NPr, we have determined the three-dimensional structure of NPr by solution NMR spectroscopy. In total, we obtained 973 NOE-derived distance restraints, 112 chemical shift-derived backbone angle restraints, and 35 hydrogen-bond restraints derived from temperature coefficients (wave). We propose that temperature wave is useful for identifying exposed beta-strands and assists in establishing protein folds based on chemical shifts. The deduced structure of NPr contains three alpha-helices and four beta-strands with the three helices all packed on the same face of the beta-sheet. The active site residue His16 of NPr for phosphoryl transfer was found to be neutral and in the N epsilon 2-H tautomeric state. There appears to be increased motion in the active site region of NPr compared to HPr, a homologous protein involved in the uptake and regulation of carbohydrate utilization.
Organizational Affiliation:
Eppley Institute for Research in Cancer and Allied Diseases, University of Nebraska Medical Center, 986805 Nebraska Medical Center, Omaha, NE 68198-6805, USA.