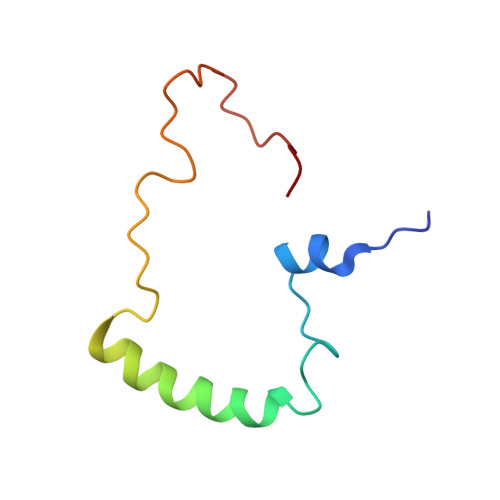
Nuclear Magnetic Resonance Structure of the Human Polyoma JC Virus Agnoprotein.
Coric, P., Saribas, A.S., Abou-Gharbia, M., Childers, W., Condra, J.H., White, M.K., Safak, M., Bouaziz, S.(2017) J Cell Biochem 118: 3268-3280
- PubMed: 28295503
- DOI: https://doi.org/10.1002/jcb.25977
- Primary Citation of Related Structures:
5NHQ - PubMed Abstract:
Agnoprotein is an important regulatory protein of the human polyoma JC virus (JCV) and plays critical roles during the viral replication cycle. It forms highly stable dimers and oligomers through its Leu/Ile/Phe-rich domain, which is important for the stability and function of the protein. We recently resolved the partial 3D structure of this protein by NMR using a synthetic peptide encompassing amino acids Thr17 to Gln52, where the Leu/Ile/Phe- rich region was found to adopt a major alpha-helix conformation spanning amino acids 23-39. Here, we report the resolution of the 3D structure of full-length JCV agnoprotein by NMR, which not only confirmed the existence of the previously reported major α-helix domain at the same position but also revealed the presence of an additional minor α-helix region spanning amino acid residues Leu6 to lys13. The remaining regions of the protein adopt an intrinsically unstructured conformation. J. Cell. Biochem. 118: 3268-3280, 2017. © 2017 Wiley Periodicals, Inc.
Organizational Affiliation:
Université Paris Descartes, Sorbonne Paris Cité, Laboratoire de Cristallographie et RMN Biologiques, UMR 8015 CNRS, 4 av. de l'Observatoire, Paris, France.