Crystal structure of Mycobacterium avium SerB2 with serine present at slightly different position near ACT domain
Shree, S., Ramachandran, R.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Phosphoserine phosphatase | 396 | Mycobacterium avium 104 | Mutation(s): 1 Gene Names: serB, MAV_3907 EC: 3.1.3.3 | 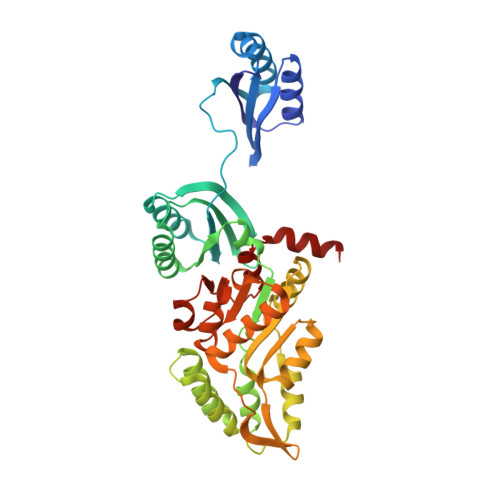 | |
UniProt | |||||
Find proteins for A0QJI1 (Mycobacterium avium (strain 104)) Explore A0QJI1 Go to UniProtKB: A0QJI1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0QJI1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PG0 Query on PG0 | D [auth A] | 2-(2-METHOXYETHOXY)ETHANOL C5 H12 O3 SBASXUCJHJRPEV-UHFFFAOYSA-N |  | ||
| SER Query on SER | E [auth A] | SERINE C3 H7 N O3 MTCFGRXMJLQNBG-REOHCLBHSA-N |  | ||
| EDO Query on EDO | C [auth A] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| MG Query on MG | B [auth A] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 67.498 | α = 90 |
| b = 107.805 | β = 90 |
| c = 133.743 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data scaling |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| PHENIX | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| CSIR | India | -- |