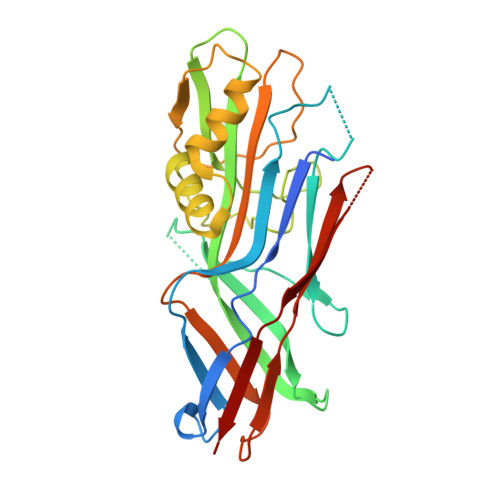
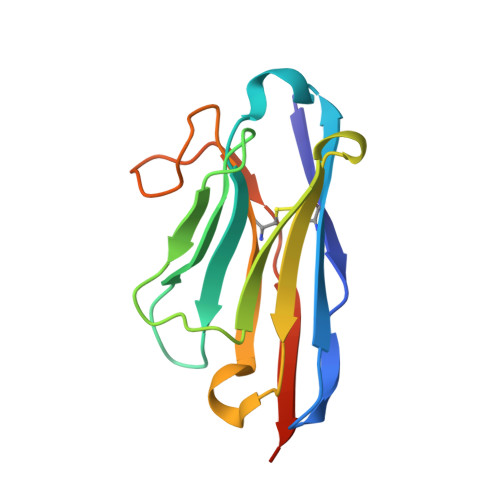
Structural insight in the inhibition of adherence of F4 fimbriae producing enterotoxigenic Escherichia coli by llama single domain antibodies.
Moonens, K., Van den Broeck, I., Okello, E., Pardon, E., De Kerpel, M., Remaut, H., De Greve, H.(2015) Vet Res 46: 14-14
- PubMed: 25828907
- DOI: https://doi.org/10.1186/s13567-015-0151-x
- Primary Citation of Related Structures:
4WEM, 4WEN, 4WEU - PubMed Abstract:
Enterotoxigenic Escherichia coli that cause neonatal and post-weaning diarrhea in piglets express F4 fimbriae to mediate attachment towards host receptors. Recently we described how llama single domain antibodies (VHHs) fused to IgA, produced in Arabidopsis thaliana seeds and fed to piglets resulted in a progressive decline in shedding of F4 positive ETEC bacteria. Here we present the structures of these inhibiting VHHs in complex with the major adhesive subunit FaeG. A conserved surface, distant from the lactose binding pocket, is targeted by these VHHs, highlighting the possibility of targeting epitopes on single-domain adhesins that are non-involved in receptor binding.