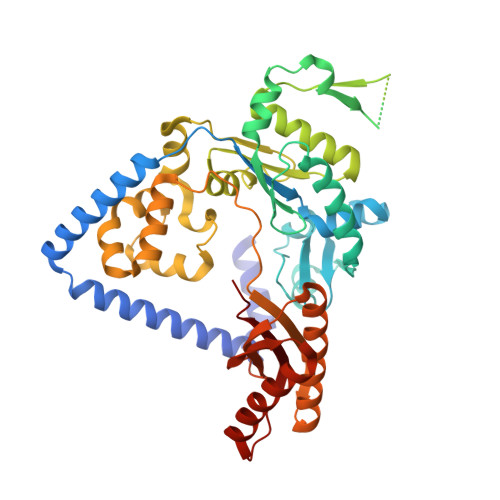
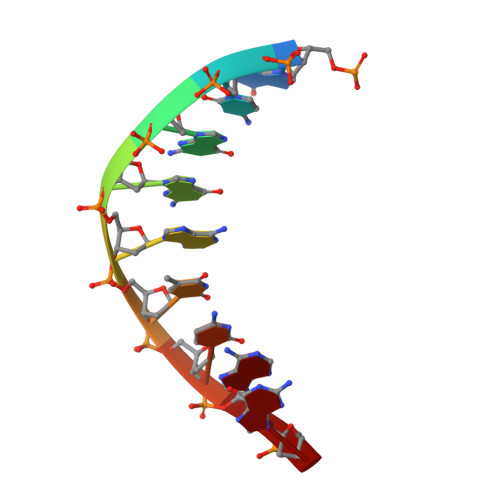
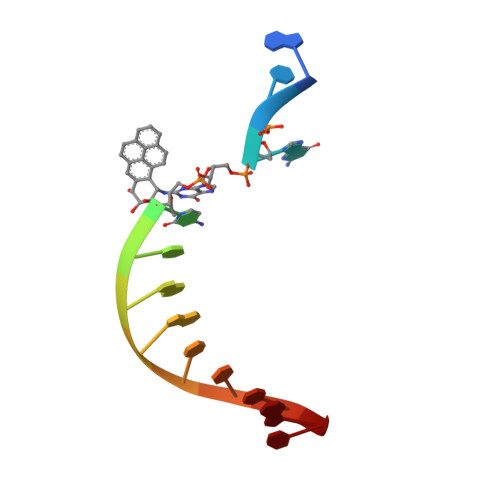
Structure and mechanism of error-free replication past the major benzo[a]pyrene adduct by human DNA polymerase kappa.
Jha, V., Bian, C., Xing, G., Ling, H.(2016) Nucleic Acids Res 44: 4957-4967
- PubMed: 27034468
- DOI: https://doi.org/10.1093/nar/gkw204
- Primary Citation of Related Structures:
4U6P, 4U7C - PubMed Abstract:
Benzo[a]pyrene (BP) is a well-known and frequently encountered carcinogen which generates a bulky DNA adduct (+)-trans-10S-BP-N(2)-dG (BP-dG) in cells. DNA polymerase kappa (polκ) is the only known Y-family polymerase that bypasses BP-dG accurately and thus protects cells from genotoxic BP. Here, we report the structures of human polκ in complex with DNA containing either a normal guanine (G) base or a BP-dG adduct at the active site and a correct deoxycytidine. The structures and supporting biochemical data reveal a unique mechanism for accurate replication by translesion synthesis past the major bulky adduct. The active site of polκ opens at the minor groove side of the DNA substrate to accommodate the bulky BP-dG that is attached there. More importantly, polκ stabilizes the lesion DNA substrate in the same active conformation as for regular B-form DNA substrates and the bulky BPDE ring in a 5' end pointing conformation. The BP-dG adducted DNA substrate maintains a Watson-Crick (BP-dG:dC) base pair within the active site, governing correct nucleotide insertion opposite the bulky adduct. In addition, polκ's unique N-clasp domain supports the open conformation of the enzyme and the extended conformation of the single-stranded template to allow bypass of the bulky lesion. This work illustrates the first molecular mechanism for how a bulky major adduct is replicated accurately without strand misalignment and mis-insertion.
Organizational Affiliation:
Department of Biochemistry, Schulich School of Medicine & Dentistry, University of Western Ontario, London, Ontario, N6A 5C1, Canada.