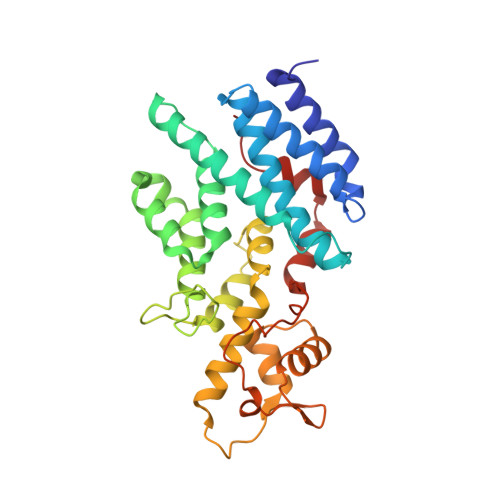
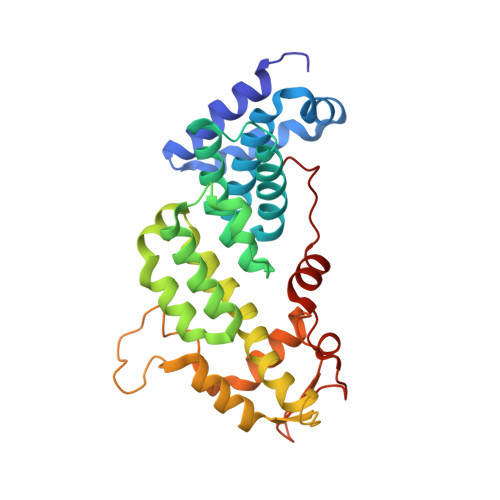
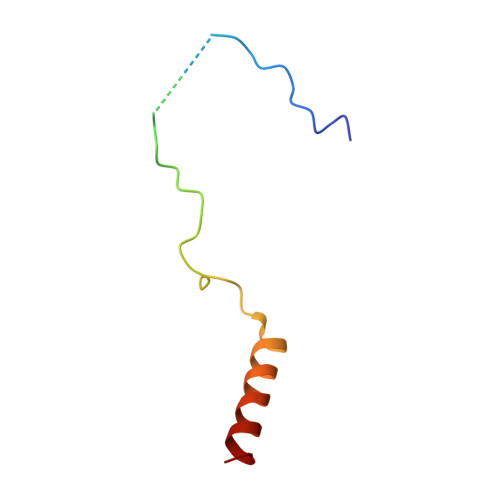
The Nuclear Pore-Associated TREX-2 Complex Employs Mediator to Regulate Gene Expression.
Schneider, M., Hellerschmied, D., Schubert, T., Amlacher, S., Vinayachandran, V., Reja, R., Pugh, B.F., Clausen, T., Kohler, A.(2015) Cell 162: 1016-1028
- PubMed: 26317468
- DOI: https://doi.org/10.1016/j.cell.2015.07.059
- Primary Citation of Related Structures:
4TRQ - PubMed Abstract:
Nuclear pore complexes (NPCs) influence gene expression besides their established function in nuclear transport. The TREX-2 complex localizes to the NPC basket and affects gene-NPC interactions, transcription, and mRNA export. How TREX-2 regulates the gene expression machinery is unknown. Here, we show that TREX-2 interacts with the Mediator complex, an essential regulator of RNA Polymerase (Pol) II. Structural and biochemical studies identify a conserved region on TREX-2, which directly binds the Mediator Med31/Med7N submodule. TREX-2 regulates assembly of Mediator with the Cdk8 kinase and is required for recruitment and site-specific phosphorylation of Pol II. Transcriptome and phenotypic profiling confirm that TREX-2 and Med31 are functionally interdependent at specific genes. TREX-2 additionally uses its Mediator-interacting surface to regulate mRNA export suggesting a mechanism for coupling transcription initiation and early steps of mRNA processing. Our data provide mechanistic insight into how an NPC-associated adaptor complex accesses the core transcription machinery.
Organizational Affiliation:
Max F. Perutz Laboratories, Medical University of Vienna, Vienna Biocenter Campus (VBC), Dr. Bohr-Gasse 9/3, 1030 Vienna, Austria.