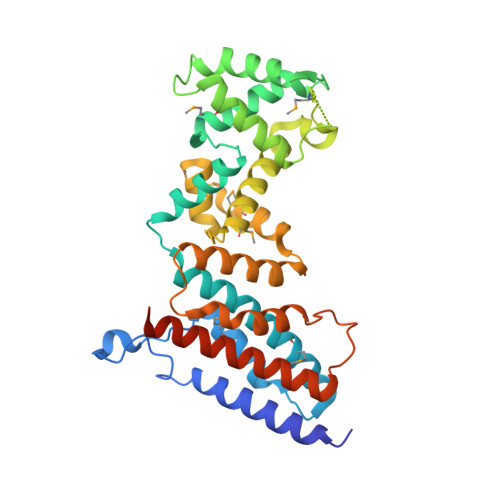
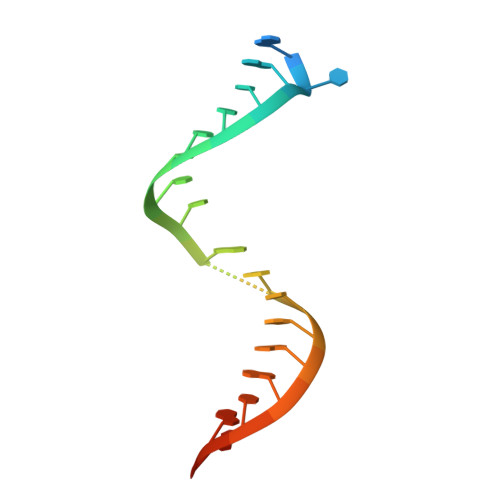
The ROQ domain of Roquin recognizes mRNA constitutive-decay element and double-stranded RNA.
Tan, D., Zhou, M., Kiledjian, M., Tong, L.(2014) Nat Struct Mol Biol 21: 679-685
- PubMed: 25026078
- DOI: https://doi.org/10.1038/nsmb.2857
- Primary Citation of Related Structures:
4QIK, 4QIL - PubMed Abstract:
A conserved stem-loop motif of the constitutive decay element (CDE) in the 3' UTR of mRNAs is recognized by the ROQ domain of Roquin, which mediates mRNA degradation. Here we report two crystal structures of the Homo sapiens ROQ domain in complex with CDE RNA. The ROQ domain has an elongated shape with three subdomains. The 19-nt Hmgxb3 CDE is bound as a stem-loop to domain III. The 23-nt TNF RNA is bound as a duplex to a separate site at the interface between domains I and II. Mutagenesis studies confirm that the ROQ domain has two separate RNA-binding sites, one for stem-loop RNA (A site) and the other for double-stranded RNA (B site). Mutation in either site perturbs the Roquin-mediated degradation of HMGXB3 and IL6 mRNAs in human cells, demonstrating the importance of both sites for mRNA decay.
Organizational Affiliation:
Department of Biological Sciences, Columbia University, New York, New York, USA.