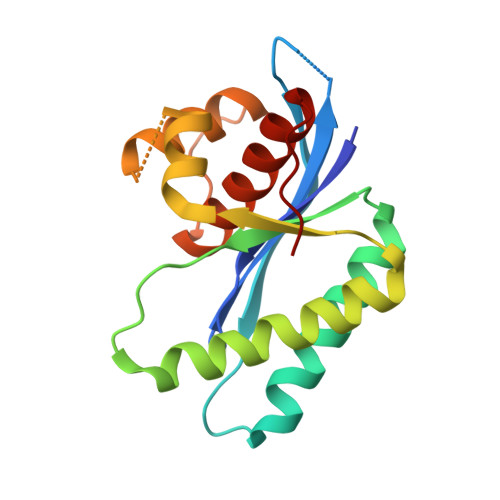
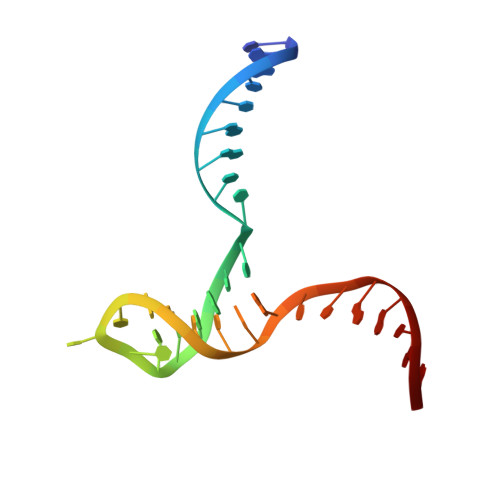
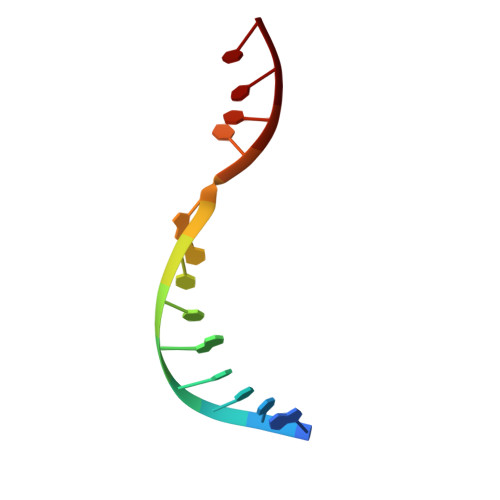
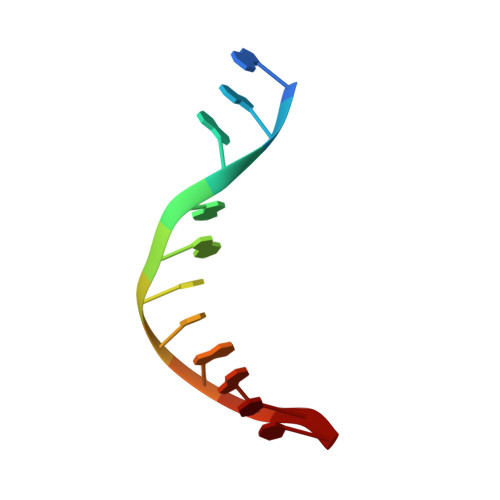
Crystal structure of RuvC resolvase in complex with Holliday junction substrate.
Gorecka, K.M., Komorowska, W., Nowotny, M.(2013) Nucleic Acids Res 41: 9945-9955
- PubMed: 23980027
- DOI: https://doi.org/10.1093/nar/gkt769
- Primary Citation of Related Structures:
4LD0 - PubMed Abstract:
The key intermediate in genetic recombination is the Holliday junction (HJ), a four-way DNA structure. At the end of recombination, HJs are cleaved by specific nucleases called resolvases. In Gram-negative bacteria, this cleavage is performed by RuvC, a dimeric endonuclease that belongs to the retroviral integrase superfamily. Here, we report the first crystal structure of RuvC in complex with a synthetic HJ solved at 3.75 Å resolution. The junction in the complex is in an unfolded 2-fold symmetrical conformation, in which the four arms point toward the vertices of a tetrahedron. The two scissile phosphates are located one nucleotide from the strand exchange point, and RuvC approaches them from the minor groove side. The key protein-DNA contacts observed in the structure were verified using a thiol-based site-specific cross-linking approach. Compared with known complex structures of the phage resolvases endonuclease I and endonuclease VII, the RuvC structure exhibits striking differences in the mode of substrate binding and location of the cleavage site.
Organizational Affiliation:
Laboratory of Protein Structure, International Institute of Molecular and Cell Biology, 4 Trojdena Street, 02-109, Warsaw, Poland.