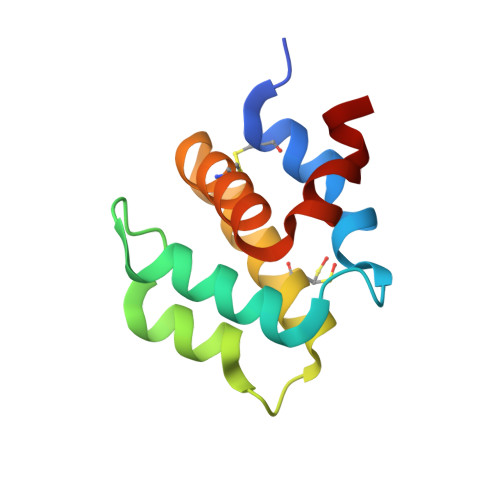
The structure of the caspase recruitment domain of BinCARD reveals that all three cysteines can be oxidized.
Chen, K.E., Richards, A.A., Caradoc-Davies, T.T., Vajjhala, P.R., Robin, G., Lua, L.H., Hill, J.M., Schroder, K., Sweet, M.J., Kellie, S., Kobe, B., Martin, J.(2013) Acta Crystallogr D Biol Crystallogr 69: 774-784
- PubMed: 23633586
- DOI: https://doi.org/10.1107/S0907444913001558
- Primary Citation of Related Structures:
4DWN, 4FH0 - PubMed Abstract:
The caspase recruitment domain (CARD) is present in death-domain superfamily proteins involved in inflammation and apoptosis. BinCARD is named for its ability to interact with Bcl10 and inhibit downstream signalling. Human BinCARD is expressed as two isoforms that encode the same N-terminal CARD region but which differ considerably in their C-termini. Both isoforms are expressed in immune cells, although BinCARD-2 is much more highly expressed. Crystals of the CARD fold common to both had low symmetry (space group P1). Molecular replacement was unsuccessful in this low-symmetry space group and, as the construct contains no methionines, first one and then two residues were engineered to methionine for MAD phasing. The double-methionine variant was produced as a selenomethionine derivative, which was crystallized and the structure was solved using data measured at two wavelengths. The crystal structures of the native and selenomethionine double mutant were refined to high resolution (1.58 and 1.40 Å resolution, respectively), revealing the presence of a cis-peptide bond between Tyr39 and Pro40. Unexpectedly, the native crystal structure revealed that all three cysteines were oxidized. The mitochondrial localization of BinCARD-2 and the susceptibility of its CARD region to redox modification points to the intriguing possibility of a redox-regulatory role.
Organizational Affiliation:
Division of Chemistry and Structural Biology, Institute for Molecular Bioscience, University of Queensland, Brisbane, Queensland 4072, Australia.