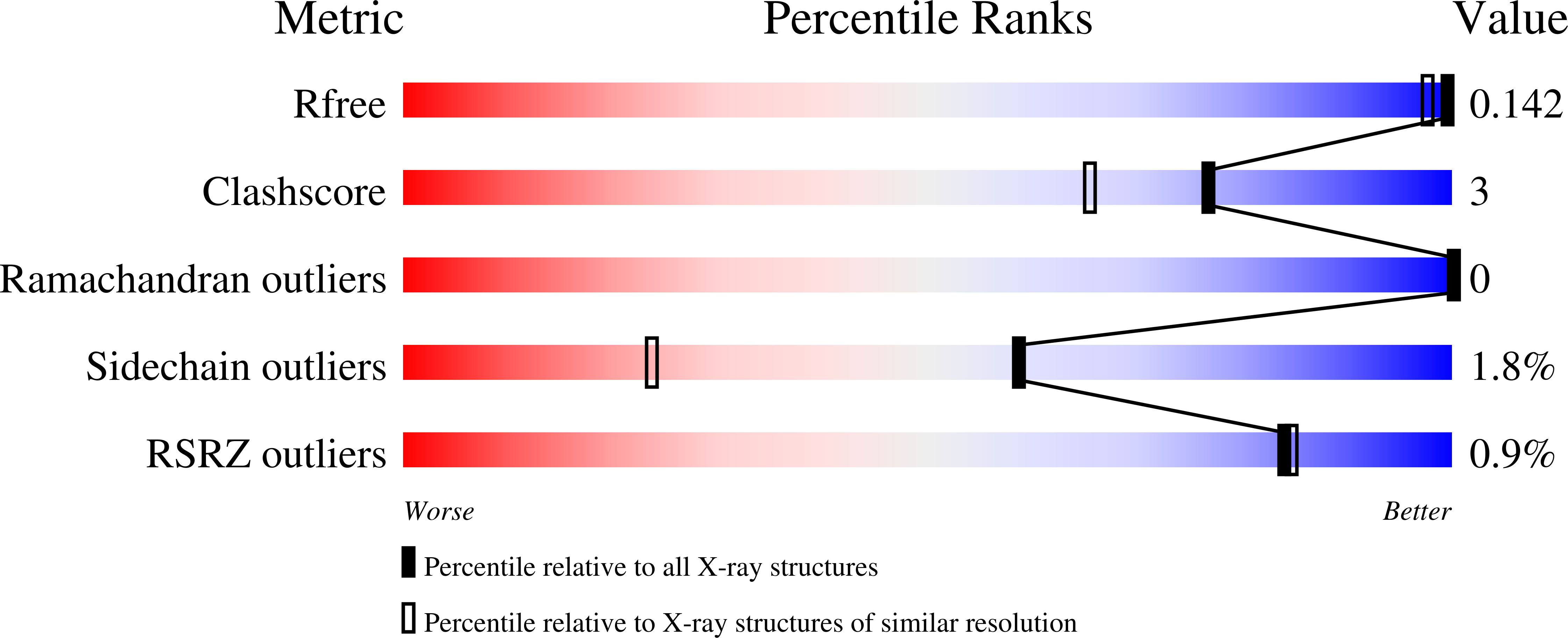
Ligand-induced fit affects binding modes and provokes changes in crystal packing of aldose reductase
Koch, C., Heine, A., Klebe, G.(2011) Biochim Biophys Acta 1810: 879-887
- PubMed: 21684320
- DOI: https://doi.org/10.1016/j.bbagen.2011.06.001
- Primary Citation of Related Structures:
3LEN, 3M0I, 3M64, 3MB9, 3MC5, 3P2V - PubMed Abstract:
Flexibility is a common feature of proteins. For human aldose reductase, a variety of conformers have been observed in crystalline complexes with different inhibitors.
Organizational Affiliation:
Philipps-Universität, Department of Pharmaceutical Chemistry, Marburg, Germany.