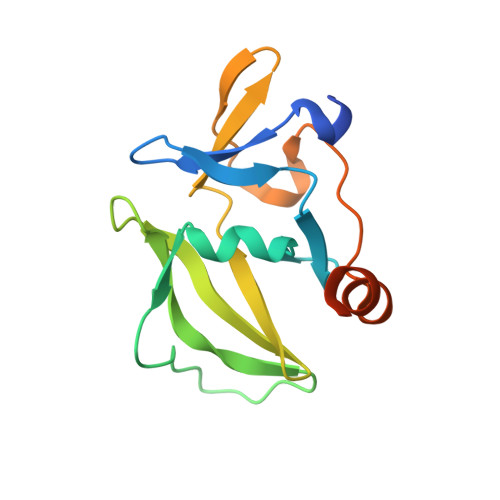
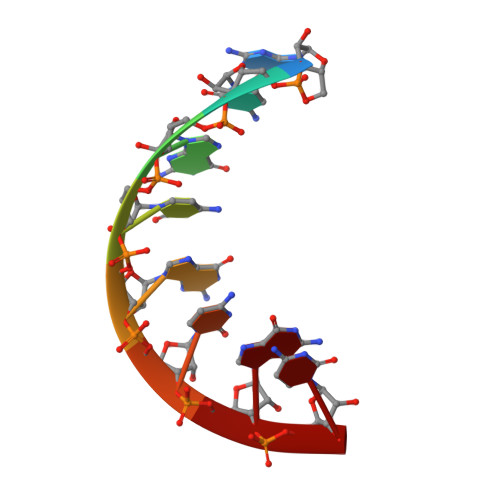
The RIG-I-like Receptor LGP2 Recognizes the Termini of Double-stranded RNA
Li, X., Ranjith-Kumar, C.T., Brooks, M.T., Dharmaiah, S., Herr, A.B., Kao, C., Li, P.(2009) J Biol Chem 284: 13881-13891
- PubMed: 19278996
- DOI: https://doi.org/10.1074/jbc.M900818200
- Primary Citation of Related Structures:
3EQT - PubMed Abstract:
The RIG-I-like receptors (RLRs), RIG-I and MDA5, recognize single-stranded RNA with 5' triphosphates and double-stranded RNA (dsRNA) to initiate innate antiviral immune responses. LGP2, a homolog of RIG-I and MDA5 that lacks signaling capability, regulates the signaling of the RLRs. To establish the structural basis of dsRNA recognition by the RLRs, we have determined the 2.0-A resolution crystal structure of human LGP2 C-terminal domain bound to an 8-bp dsRNA. Two LGP2 C-terminal domain molecules bind to the termini of dsRNA with minimal contacts between the protein molecules. Gel filtration chromatography and analytical ultracentrifugation demonstrated that LGP2 binds blunt-ended dsRNA of different lengths, forming complexes with 2:1 stoichiometry. dsRNA with protruding termini bind LGP2 and RIG-I weakly and do not stimulate the activation of RIG-I efficiently in cells. Surprisingly, full-length LGP2 containing mutations that abolish dsRNA binding retained the ability to inhibit RIG-I signaling.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Texas A & M University, College Station, Texas 77843-2128.