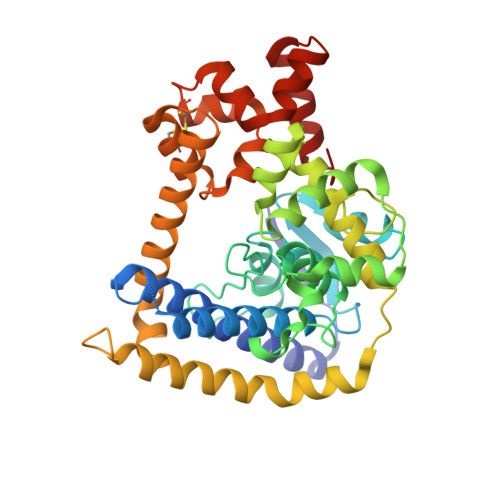
Structures of minimal catalytic fragments of topoisomerase v reveals conformational changes relevant for DNA binding.
Rajan, R., Taneja, B., Mondragon, A.(2010) Structure 18: 829-838
- PubMed: 20637419
- DOI: https://doi.org/10.1016/j.str.2010.03.006
- Primary Citation of Related Structures:
3M6K, 3M6Z, 3M7D, 3M7G - PubMed Abstract:
Topoisomerase V is an archaeal type I topoisomerase that is unique among topoisomerases due to presence of both topoisomerase and DNA repair activities in the same protein. It is organized as an N-terminal topoisomerase domain followed by 24 tandem helix-hairpin-helix (HhH) motifs. Structural studies have shown that the active site is buried by the (HhH) motifs. Here we show that the N-terminal domain can relax DNA in the absence of any HhH motifs and that the HhH motifs are required for stable protein-DNA complex formation. Crystal structures of various topoisomerase V fragments show changes in the relative orientation of the domains mediated by a long bent linker helix, and these movements are essential for the DNA to enter the active site. Phosphate ions bound to the protein near the active site helped model DNA in the topoisomerase domain and show how topoisomerase V may interact with DNA.
Organizational Affiliation:
Department of Biochemistry, Molecular Biology and Cell Biology, Northwestern University, 2205 Tech Drive, Evanston, IL 60208, USA.