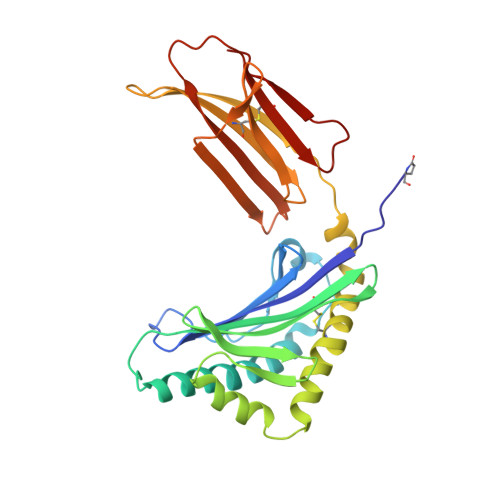
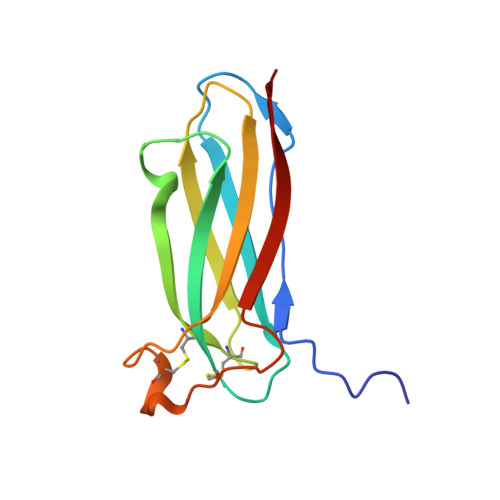
Crystal structure of the novel complex formed between zinc alpha2-glycoprotein (ZAG) and prolactin-inducible protein (PIP) from human seminal plasma
Hassan, M.I., Bilgrami, S., Kumar, V., Singh, N., Yadav, S., Kaur, P., Singh, T.P.(2008) J Mol Biol 384: 663-672
- PubMed: 18930737
- DOI: https://doi.org/10.1016/j.jmb.2008.09.072
- Primary Citation of Related Structures:
3ES6 - PubMed Abstract:
This is the first report on the formation of a complex between zinc alpha2-glycoprotein (ZAG) and prolactin-inducible protein (PIP). The complex was purified from human seminal plasma and crystallized using 20% polyethylene glycol 9000 and 5% hexaethylene glycol. The structure of the complex has been determined using X-ray crystallographic method and refined to an R(cryst) of 0.199 (R(free)=0.239). The structure of ZAG is broadly similar to the structure of serum ZAG. The scaffolding of PIP consists of seven beta-strands that are organized in the form of two antiparallel beta-pleated sheets, resulting in the formation of a sandwiched beta-sheet. The amino acid sequence of PIP contains one potential N-glycosylation site at Asn77, and the same is found glycosylated with four sugar residues. The structure of the complex shows that the beta-structure of PIP is ideally aligned with the beta-structure of domain alpha3 of ZAG to form a long interface between two proteins. The proximal beta-strands at the long interface are arranged in an antiparallel manner. There are 12 hydrogen bonds and three salt bridges between ZAG and PIP. At the two ends of vertical interface, two salt bridges are formed between pairs of Lys41-Asp233 and Lys68-Glu229. On the perpendicular interface involving alpha1-alpha2 domains of ZAG and a loop of PIP, another salt bridge is formed. The internal space at the corner of the L-shaped structure is filled with solvent molecules including a carbonate ion. The overall buried area in the complex is approximately 914 A(2), which is considerably higher than the 660 A(2) reported for the class I major histocompatibility complex structures.
Organizational Affiliation:
Department of Biophysics, All India Institute of Medical Sciences, New Delhi 110029, India.