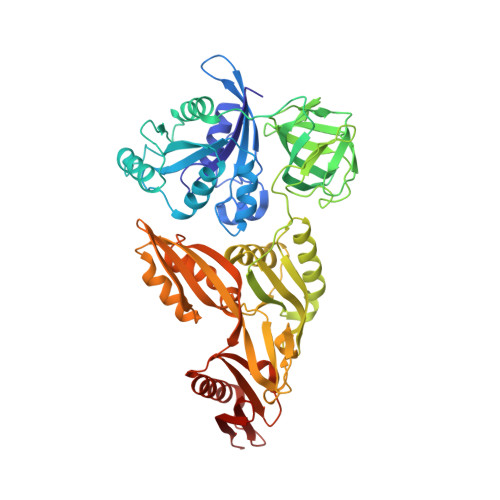
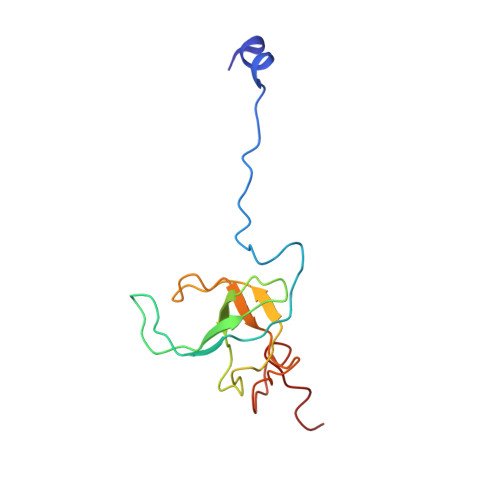
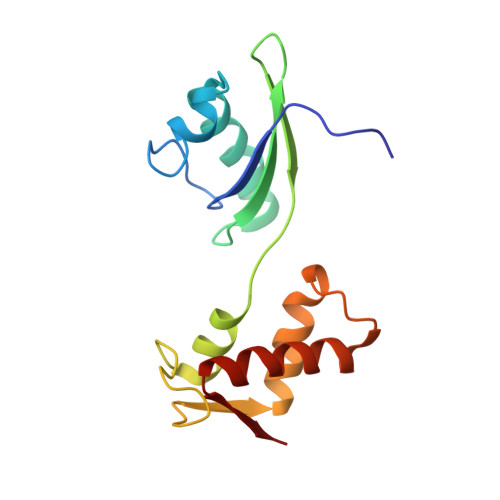
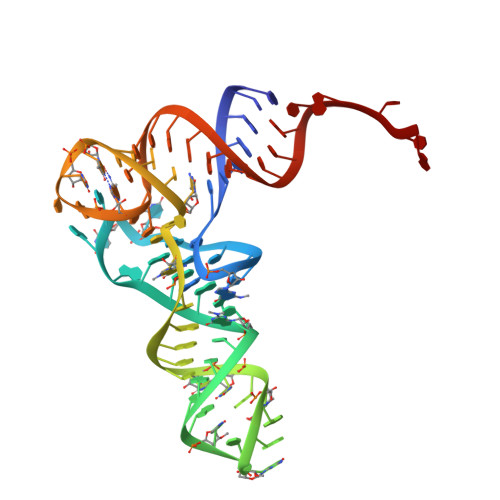
A new tRNA intermediate revealed on the ribosome during EF4-mediated back-translocation
Connell, S.R., Topf, M., Qin, Y., Wilson, D.N., Mielke, T., Fucini, P., Nierhaus, K.H., Spahn, C.M.T.(2008) Nat Struct Mol Biol 15: 910-915
- PubMed: 19172743
- DOI: https://doi.org/10.1038/nsmb.1469
- Primary Citation of Related Structures:
3DEG - PubMed Abstract:
EF4 (LepA) is an almost universally conserved translational GTPase in eubacteria. It seems to be essential under environmental stress conditions and has previously been shown to back-translocate the tRNAs on the ribosome, thereby reverting the canonical translocation reaction. In the current work, EF4 was directly visualized in the process of back-translocating tRNAs by single-particle cryo-EM. Using flexible fitting methods, we built a model of ribosome-bound EF4 based on the cryo-EM map and a recently published unbound EF4 X-ray structure. The cryo-EM map establishes EF4 as a noncanonical elongation factor that interacts not only with the elongating ribosome, but also with the back-translocated tRNA in the A-site region, which is present in a previously unseen, intermediate state and deviates markedly from the position of a canonical A-tRNA. Our results, therefore, provide insight into the underlying structural principles governing back-translocation.
Organizational Affiliation:
Institut für Medizinische Physik und Biophysik, Charite-Universitätsmedizin Berlin, Ziegelstrasse 5-9, 10117-Berlin, Germany.