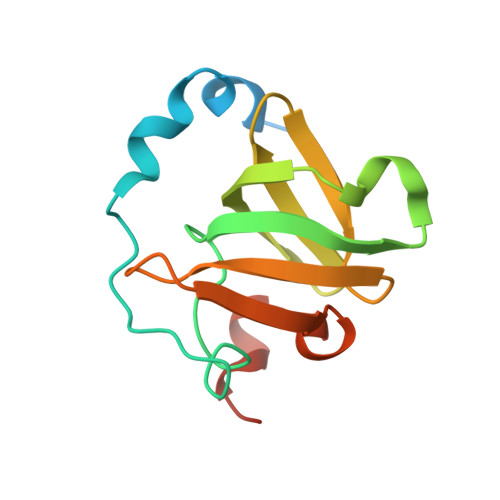
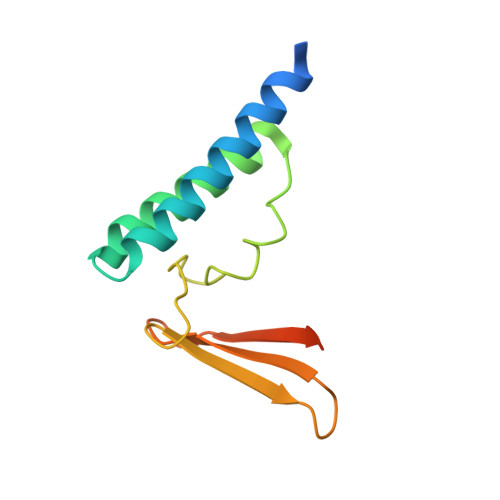
Structure of an archaeal homolog of the human protein complex Rpp21-Rpp29 that is a key core component for the assembly of active ribonuclease P.
Honda, T., Kakuta, Y., Kimura, K., Saho, J., Kimura, M.(2008) J Mol Biol 384: 652-662
- PubMed: 18929577
- DOI: https://doi.org/10.1016/j.jmb.2008.09.056
- Primary Citation of Related Structures:
2ZAE - PubMed Abstract:
Ribonuclease P (RNase P) is a ribonucleoprotein complex involved in the processing of the 5'-leader sequence of precursor tRNA. Human RNase P protein subunits Rpp21 and Rpp29, which bind to each other, with catalytic RNA (H1 RNA) are sufficient for activating endonucleolytic cleavage of precursor tRNA. Here we have determined the crystal structure of the complex between the Pyrococcus horikoshii RNase P proteins PhoRpp21 and PhoRpp29, the archaeal homologs of Rpp21 and Rpp29, respectively. PhoRpp21 and PhoRpp29 form a heterodimeric structure where the two N-terminal helices (alpha1 and alpha2) in PhoRpp21 predominantly interact with the N-terminal extended structure, the beta-strand (beta2), and the C-terminal helix (alpha3) in PhoRpp29. The interface is dominated by hydrogen bonds and several salt bridges, rather than hydrophobic interactions. The electrostatic potential on the surface of the heterodimer shows a positively charged cluster on one face, suggesting a possible RNA-binding surface of the PhoRpp21-PhoRpp29 complex. The present structure, along with the result of a mutational analysis, suggests that heterodimerization between PhoRpp21 and PhoRpp29 plays an important role in the function of P. horikoshii RNase P.
Organizational Affiliation:
Laboratory of Structural Biology, Graduate School of Systems Life Sciences, Kyushu University, Hakozaki 6-10-1, Fukuoka 812-8581, Japan.