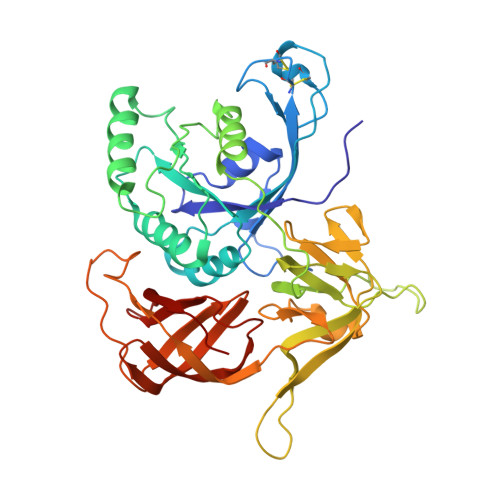
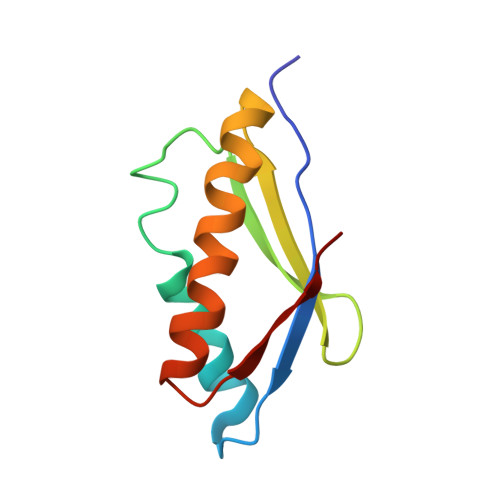
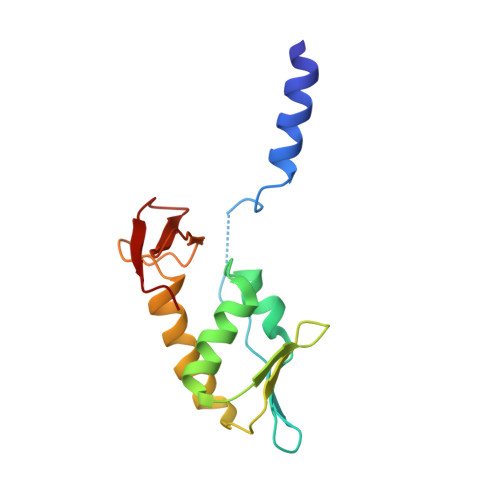
Structure of an archaeal heterotrimeric initiation factor 2 reveals a nucleotide state between the GTP and the GDP states.
Yatime, L., Mechulam, Y., Blanquet, S., Schmitt, E.(2007) Proc Natl Acad Sci U S A 104: 18445-18450
- PubMed: 18000047
- DOI: https://doi.org/10.1073/pnas.0706784104
- Primary Citation of Related Structures:
2QMU, 2QN6 - PubMed Abstract:
Initiation of translation in eukaryotes and in archaea involves eukaryotic/archaeal initiation factor (e/aIF)1 and the heterotrimeric initiation factor e/aIF2. In its GTP-bound form, e/aIF2 provides the initiation complex with Met-tRNA(i)(Met). After recognition of the start codon by initiator tRNA, e/aIF1 leaves the complex. Finally, e/aIF2, now in a GDP-bound form, loses affinity for Met-tRNA(i)(Met) and dissociates from the ribosome. Here, we report a 3D structure of an aIF2 heterotrimer from the archeon Sulfolobus solfataricus obtained in the presence of GDP. Our report highlights how the two-switch regions involved in formation of the tRNA-binding site on subunit gamma exchange conformational information with alpha and beta. The zinc-binding domain of beta lies close to the guanine nucleotide and directly contacts the switch 1 region. As a result, switch 1 adopts a not yet described conformation. Moreover, unexpectedly for a GDP-bound state, switch 2 has the "ON" conformation. The stability of these conformations is accounted for by a ligand, most probably a phosphate ion, bound near the nucleotide binding site. The structure suggests that this GDP-inorganic phosphate (Pi) bound state of aIF2 may be proficient for tRNA binding. Recently, it has been proposed that dissociation of eIF2 from the initiation complex is closely coupled to that of Pi from eIF2gamma upon start codon recognition. The nucleotide state of aIF2 shown here is indicative of a similar mechanism in archaea. Finally, we consider the possibility that release of Pi takes place after e/aIF2gamma has been informed of e/aIF1 dissociation by e/aIF2beta.
Organizational Affiliation:
Laboratoire de Biochimie, Ecole Polytechnique, Centre National de la Recherche Scientifique, F-91128 Palaiseau Cedex, France.