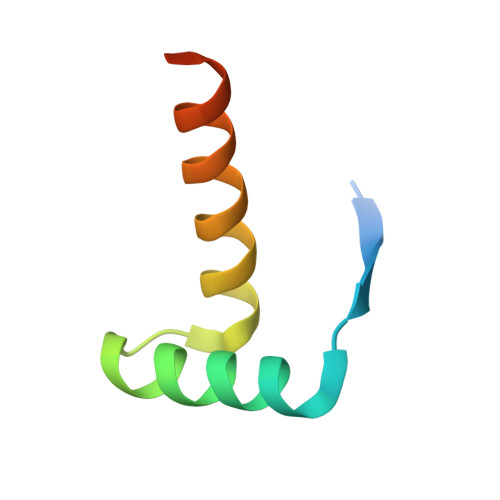
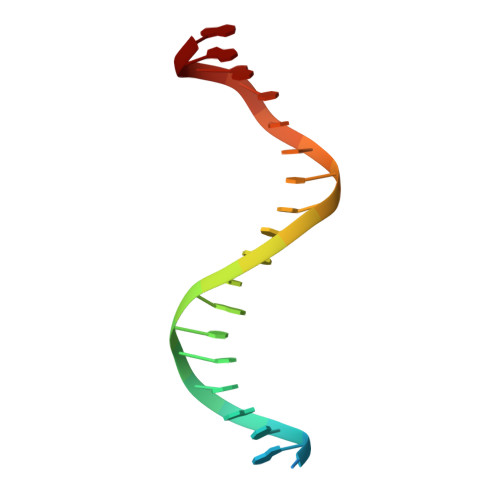
Structural basis of the transcriptional regulation of the proline utilization regulon by multifunctional PutA.
Zhou, Y., Larson, J.D., Bottoms, C.A., Arturo, E.C., Henzl, M.T., Jenkins, J.L., Nix, J.C., Becker, D.F., Tanner, J.J.(2008) J Mol Biol 381: 174-188
- PubMed: 18586269
- DOI: https://doi.org/10.1016/j.jmb.2008.05.084
- Primary Citation of Related Structures:
2RBF - PubMed Abstract:
The multifunctional Escherichia coli proline utilization A (PutA) flavoprotein functions both as a membrane-associated proline catabolic enzyme and as a transcriptional repressor of the proline utilization genes putA and putP. To better understand the mechanism of transcriptional regulation by PutA, we have mapped the put-regulatory region, determined a crystal structure of the PutA ribbon-helix-helix domain (PutA52, a polypeptide corresponding to residues 1-52 of E. coli PutA) complexed with DNA, and examined the thermodynamics of DNA binding to PutA52. Five operator sites, each containing the sequence motif 5'-GTTGCA-3', were identified using gel-shift analysis. Three of the sites are shown to be critical for repression of putA, whereas the two other sites are important for repression of putP. The 2.25-A-resolution crystal structure of PutA52 bound to one of the operators (operator 2; 21 bp) shows that the protein contacts a 9-bp fragment corresponding to the GTTGCA consensus motif plus three flanking base pairs. Since the operator sequences differ in flanking bases, the structure implies that PutA may have different affinities for the five operators. This hypothesis was explored using isothermal titration calorimetry. The binding of PutA52 to operator 2 is exothermic, with an enthalpy of -1.8 kcal/mol and a dissociation constant of 210 nM. Substitution of the flanking bases of operator 4 into operator 2 results in an unfavorable enthalpy of 0.2 kcal/mol and a 15-fold-lower affinity, showing that base pairs outside of the consensus motif impact binding. Structural and thermodynamic data suggest that hydrogen bonds between Lys9 and bases adjacent to the GTTGCA motif contribute to transcriptional regulation by fine-tuning the affinity of PutA for put control operators.
Organizational Affiliation:
Department of Biochemistry, University of Nebraska-Lincoln, Lincoln, NE 68588, USA.