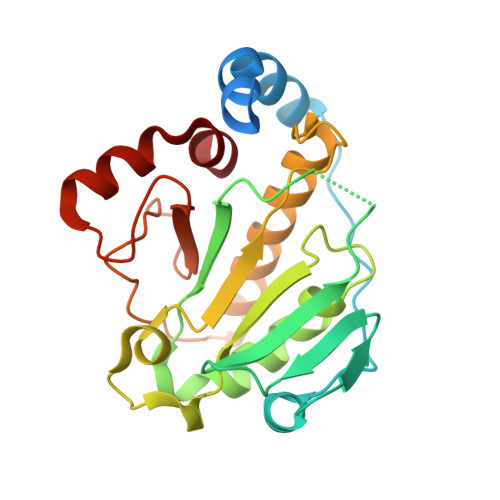
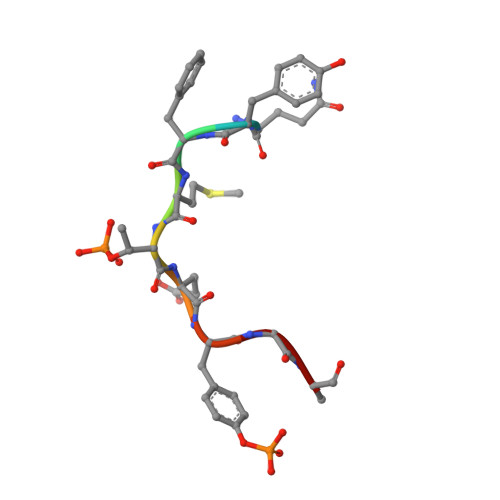
Structural insights into the enzymatic mechanism of the pathogenic MAPK phosphothreonine lyase
Zhu, Y., Li, H., Long, C., Hu, L., Xu, H., Liu, L., Chen, S., Wang, D.C., Shao, F.(2007) Mol Cell 28: 899-913
- PubMed: 18060821
- DOI: https://doi.org/10.1016/j.molcel.2007.11.011
- Primary Citation of Related Structures:
2P1W, 2Q8Y - PubMed Abstract:
The OspF family of phosphothreonine lyase, including SpvC from Salmonella, irreversibly inactivates the dual-phosphorylated host MAPKs (pT-X-pY) through beta elimination. We determined crystal structures of SpvC and its complex with a phosphopeptide substrate. SpvC adopts a unique fold of alpha/beta type. The disordered N terminus harbors a canonical D motif for MAPK substrate docking. The enzyme-substrate complex structure indicates that recognition of the phosphotyrosine followed by insertion of the threonine phosphate into an arginine pocket places the phosphothreonine into the enzyme active site. This requires the conformational flexibility of pT-X-pY, which suggests that p38 (pT-G-pY) is likely the preferred physiological substrate. Structure-based biochemical and enzymatic analysis allows us to propose a general acid/base mechanism for beta elimination reaction catalyzed by the phosphothreonine lyase. The mechanism described here provides a structural understanding of MAPK inactivation by a family of pathogenic effectors conserved in plant and animal systems and may also open a new route for biological catalysis.
Organizational Affiliation:
National Institute of Biological Sciences, Beijing, 102206, China.