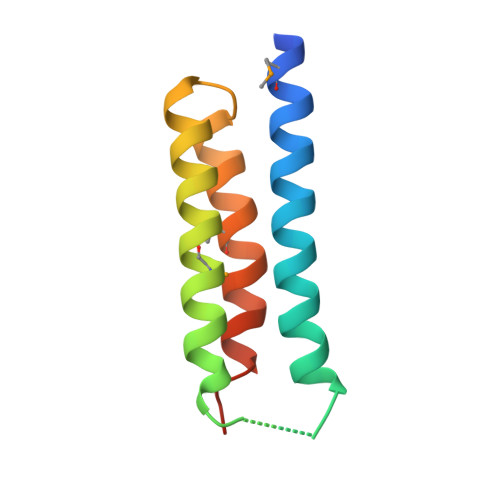
Crystal structure of S. aureus YlaN, an essential leucine rich protein involved in the control of cell shape.
Xu, L., Sedelnikova, S.E., Baker, P.J., Hunt, A., Errington, J., Rice, D.W.(2007) Proteins 68: 438-455
- PubMed: 17469204
- DOI: https://doi.org/10.1002/prot.21377
- Primary Citation of Related Structures:
2ODM - PubMed Abstract:
The crystal structure of a conserved leucine rich protein, YlaN, from Staphylococcus aureus has been determined by X-ray crystallography to 2.3 A resolution. Whilst the precise function of S. aureus YlaN is unknown its homologue in B. subtilis has been shown to be essential for cell survival and is thought to be involved in controlling cell shape. The structure of S. aureus YlaN provides the first view of its protein family, which reveals that it is a novel homodimer whose subunit architecture is comprised of an antiparallel 3 helix bundle reminiscent of the helical arrangements seen in leucine zipper proteins. Analysis of the pattern of sequence conservation on the structure has led to the identification of two connected solvent exposed patches of conserved residues in each subunit located at one end of but on opposite faces of the molecule. We suggest that YlaN has a binding role in the cell rather than a catalytic function and a search for its ligand is underway to accelerate its exploitation as a target for antibiotic discovery.
Organizational Affiliation:
Department of Molecular Biology and Biotechnology, Krebs Institute for Biomolecular Research, The University of Sheffield, Sheffield, United Kingdom.