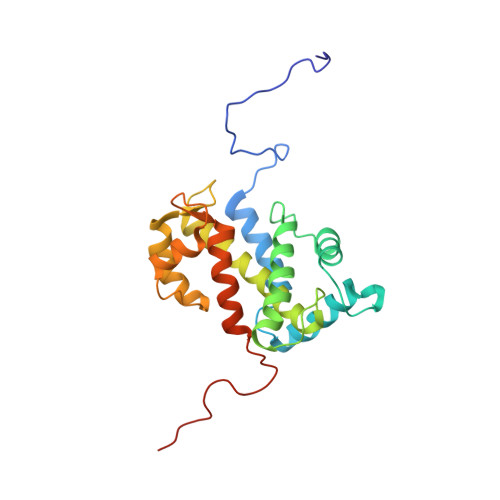
NMR structure of the calflagin Tb24 flagellar calcium binding protein of Trypanosoma brucei.
Xu, X., Olson, C.L., Engman, D.M., Ames, J.B.(2012) Protein Sci 21: 1942-1947
- PubMed: 23011904
- DOI: https://doi.org/10.1002/pro.2167
- Primary Citation of Related Structures:
2LVV - PubMed Abstract:
Flagellar calcium binding proteins are expressed in a variety of trypanosomes and are potential drug targets for Chagas disease and African sleeping sickness. The flagellar calcium binding protein calflagin of Trypanosoma brucei (called Tb24) is a myristoylated and palmitoylated EF-hand protein that is targeted to the inner leaflet of the flagellar membrane. The Tb24 protein may also interact with proteins on the membrane surface that may be different from those bound to flagellar calcium binding proteins (FCaBPs) in T. cruzi. We report here the NMR structure of Tb24 that contains four EF-hand motifs bundled in a compact arrangement, similar to the overall fold of T. cruzi FCaBP (RMSD = 1.0 Å). A cluster of basic residues (K22, K25, K31, R36, and R38) located on a surface near the N-terminal myristoyl group may be important for membrane binding. Non-conserved residues on the surface of a hydrophobic groove formed by EF2 (P91, Q95, D103, and V108) and EF4 (C194, T198, K199, Q202, and V203) may serve as a target protein binding site and could have implications for membrane target recognition.
Organizational Affiliation:
Department of Chemistry, University of California, Davis, California 95616, USA.