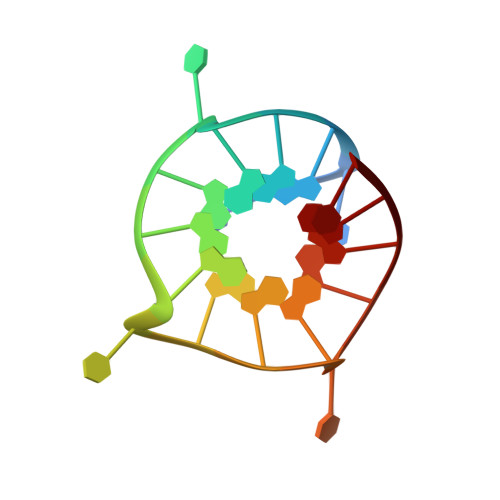
Monomer-Dimer Equilibrium for the 5'-5' Stacking of Propeller-Type Parallel-Stranded G-Quadruplexes: NMR Structural Study
Do, N.Q., Phan, A.T.(2012) Chemistry
- PubMed: 23019076
- DOI: https://doi.org/10.1002/chem.201103295
- Primary Citation of Related Structures:
2LK7 - PubMed Abstract:
Guanine-rich sequence motifs, which contain tracts of three consecutive guanines connected by single non-guanine nucleotides, are abundant in the human genome and can form a robust G-quadruplex structure with high stability. Herein, by using NMR spectroscopy, we investigate the equilibrium between monomeric and 5'-5' stacked dimeric propeller-type G-quadruplexes that are formed by DNA sequences containing GGGT motifs. We show that the monomer-dimer equilibrium depends on a number of parameters, including the DNA concentration, DNA flanking sequences, the concentration and type of cations, and the temperature. We report on the high-definition structure of a simple monomeric G-quadruplex containing three single-residue loops, which could serve as a reference for propeller-type G-quadruplex structures in solution.
Organizational Affiliation:
School of Physical and Mathematical Sciences, Nanyang Technological University, Singapore.