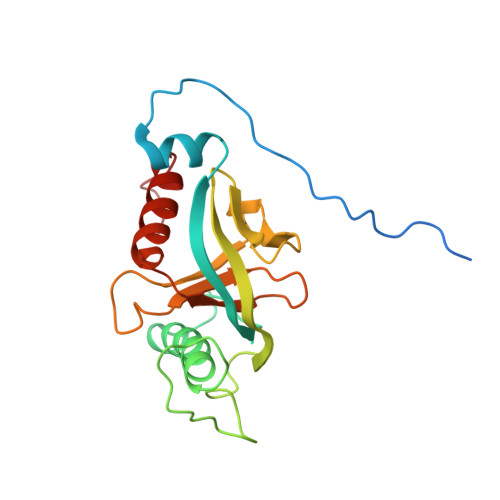
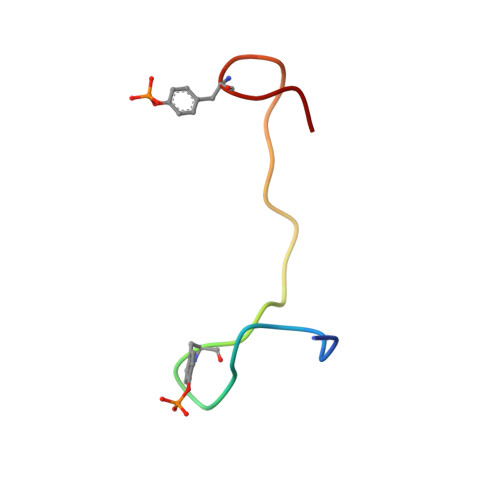
Integrin {beta}3 phosphorylation dictates its complex with the Shc phosphotyrosine-binding (PTB) domain.
Deshmukh, L., Gorbatyuk, V., Vinogradova, O.(2010) J Biol Chem 285: 34875-34884
- PubMed: 20739287
- DOI: https://doi.org/10.1074/jbc.M110.159087
- Primary Citation of Related Structures:
2L1C - PubMed Abstract:
Adaptor protein Shc plays a key role in mitogen-activated protein kinase (MAPK) signaling pathway, which can be mediated through a number of different receptors including integrins. By specifically recognizing the tyrosine-phosphorylated integrin β(3), Shc has been shown to trigger integrin outside-in signaling, although the structural basis of this interaction remains nebulous. Here we present the detailed structural analysis of Shc phosphotyrosine-binding (PTB) domain in complex with the bi-phosphorylated β(3)integrin cytoplasmic tail (CT). We show that this complex is primarily defined by the phosphorylation state of the integrin C-terminal Tyr(759), which fits neatly into the classical PTB pocket of Shc. In addition, we have identified a novel binding interface which concurrently accommodates phosphorylated Tyr(747) of the highly conserved NPXY motif of β(3). The structure represents the first snapshot of an integrin cytoplasmic tail bound to a target for mediating the outside-in signaling. Detailed comparison with the known Shc PTB structure bound to a target TrkA peptide revealed some significant differences, which shed new light upon the PTB domain specificity.
Organizational Affiliation:
Department of Pharmaceutical Sciences, School of Pharmacy, University of Connecticut, Storrs, Connecticut 06269-3092, USA.