Promiscuous mismatch extension by human DNA polymerase lambda.
Picher, A.J., Garcia-Diaz, M., Bebenek, K., Pedersen, L.C., Kunkel, T.A., Blanco, L.(2006) Nucleic Acids Res 34: 3259-3266
- PubMed: 16807316
- DOI: https://doi.org/10.1093/nar/gkl377
- Primary Citation of Related Structures:
2GWS - PubMed Abstract:
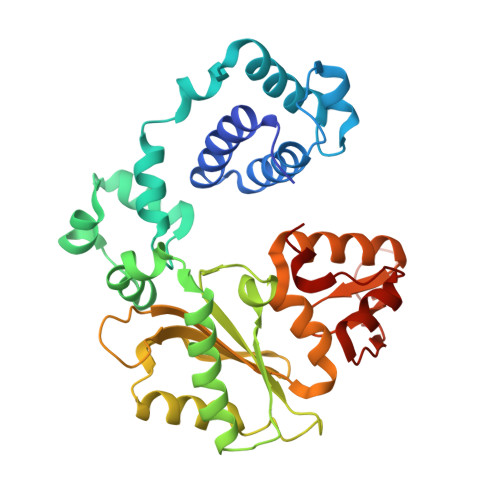
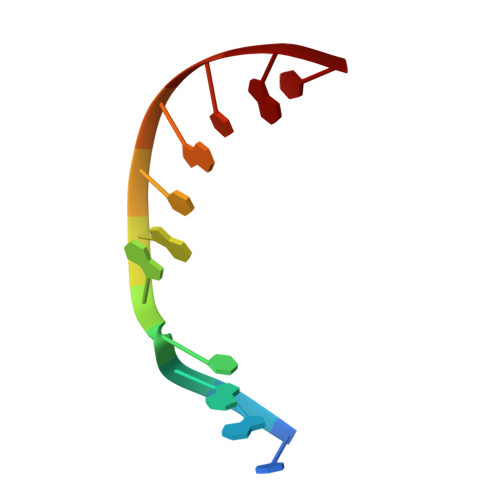
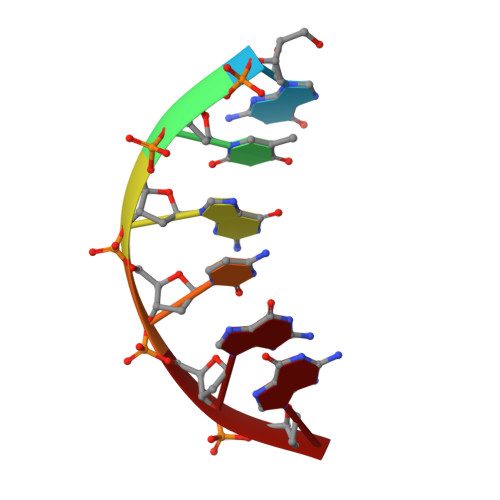
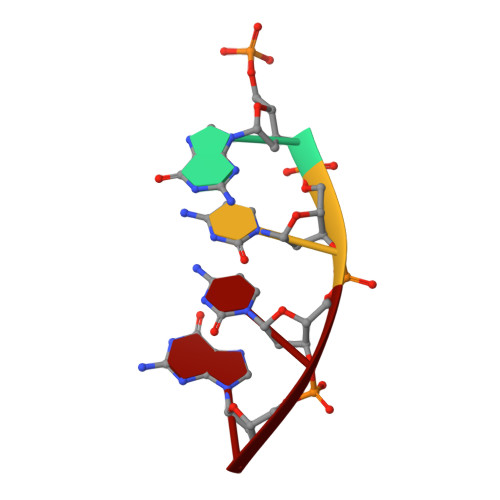
DNA polymerase lambda (Pol lambda) is one of several DNA polymerases suggested to participate in base excision repair (BER), in repair of broken DNA ends and in translesion synthesis. It has been proposed that the nature of the DNA intermediates partly determines which polymerase is used for a particular repair reaction. To test this hypothesis, here we examine the ability of human Pol lambda to extend mismatched primer-termini, either on 'open' template-primer substrates, or on its preferred substrate, a 1 nt gapped-DNA molecule having a 5'-phosphate. Interestingly, Pol lambda extended mismatches with an average efficiency of approximately 10(-2) relative to matched base pairs. The match and mismatch extension catalytic efficiencies obtained on gapped molecules were approximately 260-fold higher than on template-primer molecules. A crystal structure of Pol lambda in complex with a single-nucleotide gap containing a dG.dGMP mismatch at the primer-terminus (2.40 A) suggests that, at least for certain mispairs, Pol lambda is unable to differentiate between matched and mismatched termini during the DNA binding step, thus accounting for the relatively high efficiency of mismatch extension. This property of Pol lambda suggests a potential role as a 'mismatch extender' during non-homologous end joining (NHEJ), and possibly during translesion synthesis.
Organizational Affiliation:
Centro de Biología Molecular Severo Ochoa (CSIC-UAM), Universidad Autónoma, 28049 Madrid, Spain.