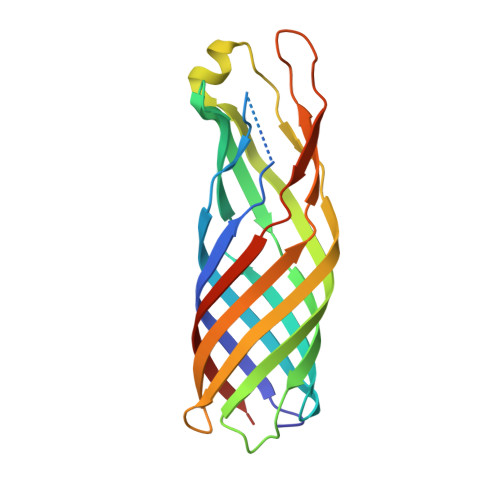
The outer membrane protein OmpW forms an eight-stranded beta-barrel with a hydrophobic channel.
Hong, H., Patel, D.R., Tamm, L.K., van den Berg, B.(2006) J Biol Chem 281: 7568-7577
- PubMed: 16414958
- DOI: https://doi.org/10.1074/jbc.M512365200
- Primary Citation of Related Structures:
2F1T, 2F1V - PubMed Abstract:
Escherichia coli OmpW belongs to a family of small outer membrane proteins that are widespread in Gram-negative bacteria. Their functions are unknown, but recent data suggest that they may be involved in the protection of bacteria against various forms of environmental stress. To gain insight into the function of these proteins A we have determined the crystal structure of E. coli OmpW to 2.7-A resolution. The structure shows that OmpW forms an 8-stranded beta-barrel with a long and narrow hydrophobic channel that contains a bound n-dodecyl-N,N-dimethylamine-N-oxide detergent molecule. Single channel conductance experiments show that OmpW functions as an ion channel in planar lipid bilayers. The channel activity can be blocked by the addition of n-dodecyl-N,N-dimethylamine-N-oxide. Taken together, the data suggest that members of the OmpW family could be involved in the transport of small hydrophobic molecules across the bacterial outer membrane.
Organizational Affiliation:
Department of Molecular Physiology and Biological Physics, University of Virginia, Charlottesville, VA 22908, USA.