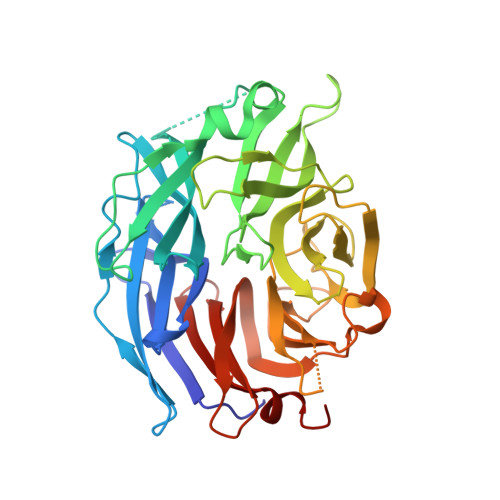
Crystal Structure of the Human Sialidase Neu2 in Complex with 2',3'- dihydroxypropyl ether mimetic Inhibitor
Chavas, L.M.G., Kato, R., Mann, M.C., Thomson, R.J., Dyason, J.C., von Itzstein, M., Fusi, P., Tringali, C., Venerando, B., Tettamanti, G., Monti, E., Wakatsuki, S.To be published.