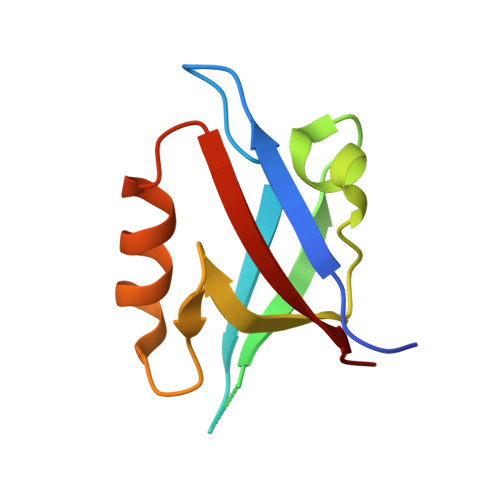
Crystal structures of autoinhibitory PDZ domain of Tamalin: implications for metabotropic glutamate receptor trafficking regulation
Sugi, T., Oyama, T., Muto, T., Nakanishi, S., Morikawa, K., Jingami, H.(2007) EMBO J 26: 2192-2205
- PubMed: 17396155
- DOI: https://doi.org/10.1038/sj.emboj.7601651
- Primary Citation of Related Structures:
2EGK, 2EGN, 2EGO - PubMed Abstract:
Metabotropic glutamate receptors (mGluRs) function as neuronal G-protein-coupled receptors and this requires efficient membrane targeting through associations with cytoplasmic proteins. However, the molecular mechanism regulating mGluR cell-surface trafficking remains unknown. We report here that mGluR trafficking is controlled by the autoregulatory assembly of a scaffold protein Tamalin. In the absence of mGluR, Tamalin self-assembles into autoinhibited conformations, through its PDZ domain and C-terminal intrinsic ligand motif. X-ray crystallographic analyses visualized integral parts of the oligomeric self-assemblies of Tamalin, which require not only the novel hydrophobic dimerization interface but also canonical and noncanonical PDZ/ligand autoinhibitory interactions. The mGluR cytoplasmic region can competitively bind to Tamalin at a higher concentration, disrupting weak inhibitory interactions. The atomic view of mGluR association suggests that this rearrangement is dominated by electrostatic attraction and repulsion. We also observed in mammalian cells that the association liberates the intrinsic ligand toward a motor protein receptor, thereby facilitating mGluR cell-surface trafficking. Our study suggests a novel regulatory mechanism of the PDZ domain, by which Tamalin switches between the trafficking-inhibited and -active forms depending on mGluR association.
Organizational Affiliation:
Department of Molecular Biology, Biomolecular Engineering Research Institute, Suita, Osaka, Japan.