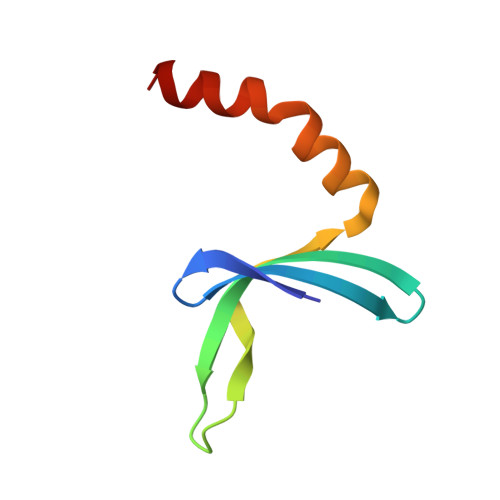
A Global Transcription Cofactor Bound to Juxtaposed Strands of Unwound DNA
Werten, S., Moras, D.(2006) Nat Struct Mol Biol 13: 181
- PubMed: 16415882
- DOI: https://doi.org/10.1038/nsmb1044
- Primary Citation of Related Structures:
2C62 - PubMed Abstract:
The 1.74-A crystal structure of the human transcription cofactor PC4 in complex with a single-stranded 20-mer oligonucleotide reveals how symmetry-related beta-surfaces of the protein homodimer interact with juxtaposed 5-nucleotide DNA regions running in opposite directions. The structure explains high-affinity binding of PC4 to the complementary strands of unwinding duplex DNA, and it suggests the cofactor may have a role in relaxing negative supercoils or exposing unpaired bases for sequence-specific recognition by other biomolecules.
Organizational Affiliation:
Département de Biologie et Génomique Structurales, Institut de Génétique et de Biologie Moléculaire et Cellulaire, B.P. 10142, 67404 Illkirch Cedex, France.