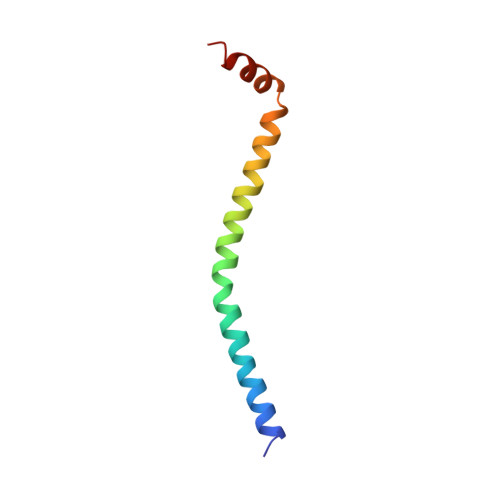
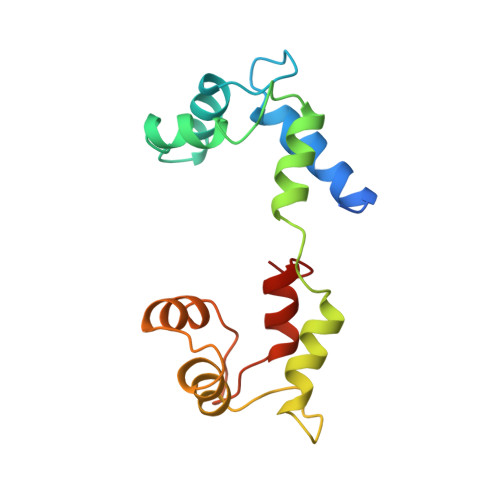
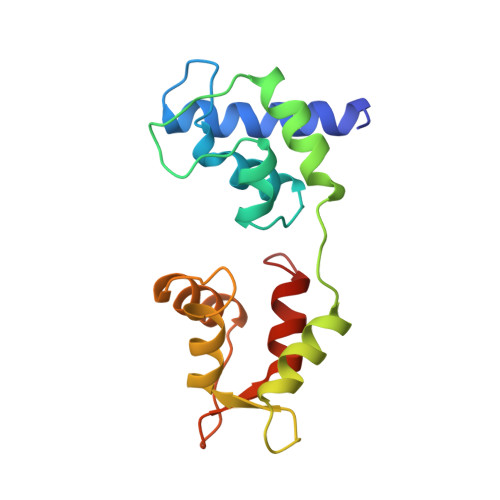
Structure of the regulatory domain of scallop myosin at 2 A resolution: implications for regulation.
Houdusse, A., Cohen, C.(1996) Structure 4: 21-32
- PubMed: 8805510
- DOI: https://doi.org/10.1016/s0969-2126(96)00006-8
- Primary Citation of Related Structures:
1WDC - PubMed Abstract:
In contrast to the myosins of vertebrate skeletal muscle, molluscan myosins are regulated molecules whose enzymatic activity is switched on by the direct binding of Ca2+. The head portion (S1) of the molecule consists of a motor domain and a regulatory domain (RD) containing a 'regulatory' and an 'essential' light chain (RLC and ELC, respectively). The structures of scallop myosin RD with bound Ca2+, as well as the S1 fragment of chicken skeletal muscle myosin, have been determined previously to 2.8 A resolution.
Organizational Affiliation:
Rosenstiel Basic Medical Sciences Research Center, Brandeis University, Waltham, MA 02254-9110, USA.