Accelerated X-ray structure elucidation of a 36 kDa muramidase/transglycosylase using wARP.
Van Asselt, E.J., Perrakis, A., Kalk, K.H., Lamzin, V.S., Dijkstra, B.W.(1998) Acta Crystallogr D Biol Crystallogr 54: 58-73
- PubMed: 9761817
- DOI: https://doi.org/10.1107/s0907444997010330
- Primary Citation of Related Structures:
1LTM - PubMed Abstract:
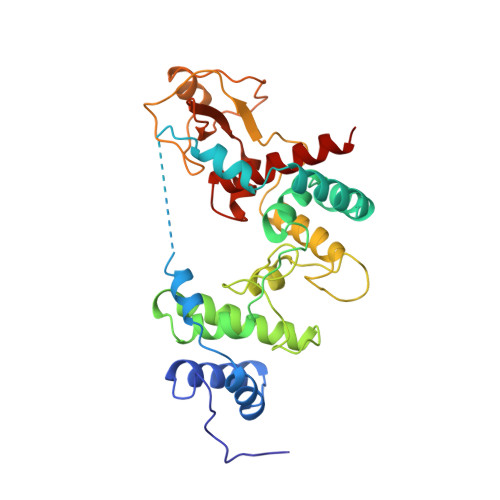
The X-ray structure of the 36 kDa soluble lytic transglycosylase from Escherichia coli has been determined starting with the multiple isomorphous replacement method with inclusion of anomalous scattering at 2.7 A resolution. Subsequently, before any model building was carried out, phases were extended to 1.7 A resolution with the weighted automated refinement procedure wARP, which gave a dramatic improvement in the phases. The electron-density maps from wARP were of outstanding quality for both the main chain and the side chains of the protein, which allowed the time spent on the tracing, interpretation and building of the X-ray structure to be substantially shortened. The structure of the soluble lytic transglycosylase was refined at 1.7 A resolution with X-PLOR to a final crystallographic R factor of 18.9%. Analysis of the wARP procedure revealed that the use of the maximum-likelihood refinement in wARP gave much better phases than least-squares refinement, provided that the ratio of reflections to protein atom parameters was approximately 1.8 or higher. Furthermore, setting aside 5% of the data for an Rfree test set had a negative effect on the phase improvement. The mean WwARP, a weight determined at the end of the wARP procedure and based on the variance of structure factors from six individually refined wARP models, proved to be a better indicator than the Rfree factor to judge different phase improvement protocols. The elongated Slt35 structure has three domains named the alpha, beta and core domains. The alpha domain contains mainly alpha-helices, while the beta domain consists of a five-stranded antiparallel beta-sheet flanked by a short alpha-helix. Sandwiched between the alpha and beta domains is the core domain, which bears some resemblance to the fold of the catalytic domain of the previously elucidated 70 kDa soluble lytic transglycosylase from E. coli. The putative active site is at the bottom of a large deep groove in the core domain.
Organizational Affiliation:
BIOSON Research Institute and Laboratory of Biophysical Chemistry, Groningen University, Nijenborgh 4, 9747 AG Groningen, The Netherlands.