The E. Coli Nusa Carboxy-Terminal Domains are Structurally Similar and Show Specific Rnap- and Lambdan Interactions
Eisenmann, A., Schwarz, S., Prasch, S., Schweimer, K., Roesch, P.(2005) Protein Sci 14: 2018
- PubMed: 15987884
- DOI: https://doi.org/10.1110/ps.051372205
- Primary Citation of Related Structures:
1WCL, 1WCN - PubMed Abstract:
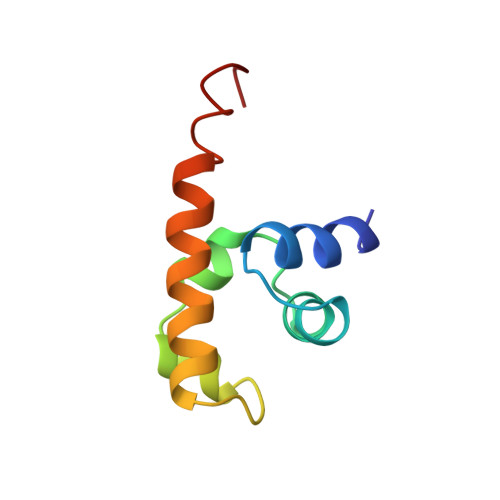
The carboxy-terminal domain of the transcription factor Escherichia coli NusA, NusACTD, interacts with the protein N of bacteriophage lambda, lambdaN, and the carboxyl terminus of the E. coli RNA polymerase alpha subunit, alphaCTD. We solved the solution structure of the unbound NusACTD with high-resolution nuclear magnetic resonance (NMR). Additionally, we investigated the binding sites of lambdaN and alphaCTD on NusACTD using NMR titrations. The solution structure of NusACTD shows two structurally similar subdomains, NusA(353-416) and NusA(431-490), matching approximately two homologous acidic sequence repeats. Further characterization of NusACTD with 15N NMR relaxation data suggests that the interdomain region is only weakly structured and that the subdomains are not interacting. Both subdomains adopt an (HhH)2 fold. These folds are normally involved in DNA-protein and protein-protein interactions. NMR titration experiments show clear differences of the interactions of these two domains with alphaCTD and lambdaN, in spite of their structural similarity.
Organizational Affiliation:
Department of Biopolymers, University of Bayreuth, 95440 Bayreuth, Germany.