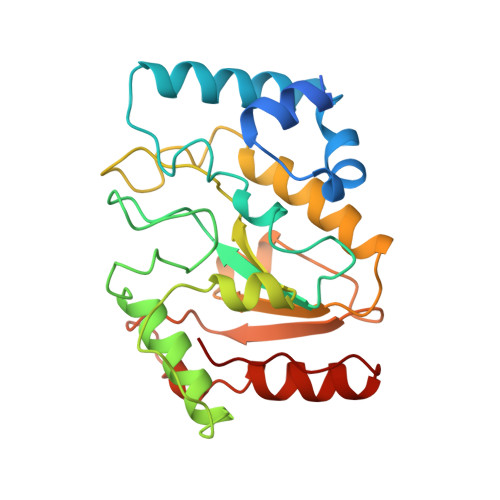
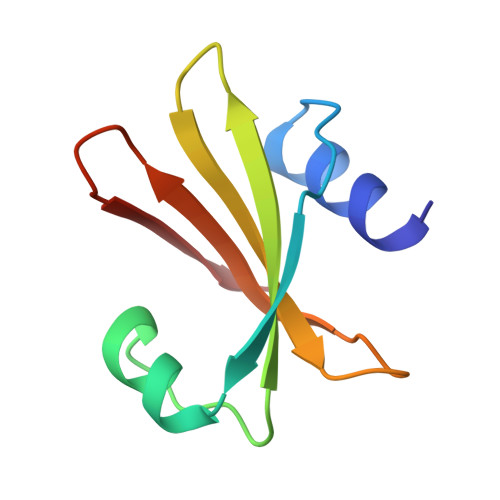
Nucleotide mimicry in the crystal structure of the uracil-DNA glycosylase-uracil glycosylase inhibitor protein complex.
Savva, R., Pearl, L.H.(1995) Nat Struct Biol 2: 752-757
- PubMed: 7552746
- DOI: https://doi.org/10.1038/nsb0995-752
- Primary Citation of Related Structures:
1UDI - PubMed Abstract:
The Bacillus subtilis bacteriophages PBS-1 and PBS-2 protect their uracil-containing DNA by expressing an inhibitor protein (UGI) which inactivates the host uracil-DNA glycosylase (UDGase) base-excision repair enzyme. Also, PBS1/2 UGI efficiently inactivates UDGases from other biological sources, including the enzyme from herpes simplex virus type-1 (HSV-1). The crystal structure of the HSV-1 UDGase-PBS1 UGI complex at 2.7 angstrum reveals an alpha-beta-alpha sandwich structure for UGI which interacts with conserved regions of UDGase involved in DNA binding, and directly mimics protein-DNA interactions observed in the UDGase-oligonucleotide complex. The inhibitor completely blocks access to the active site of UDGase, but makes no direct contact with the uracil-binding pocket itself.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University College, London, UK.